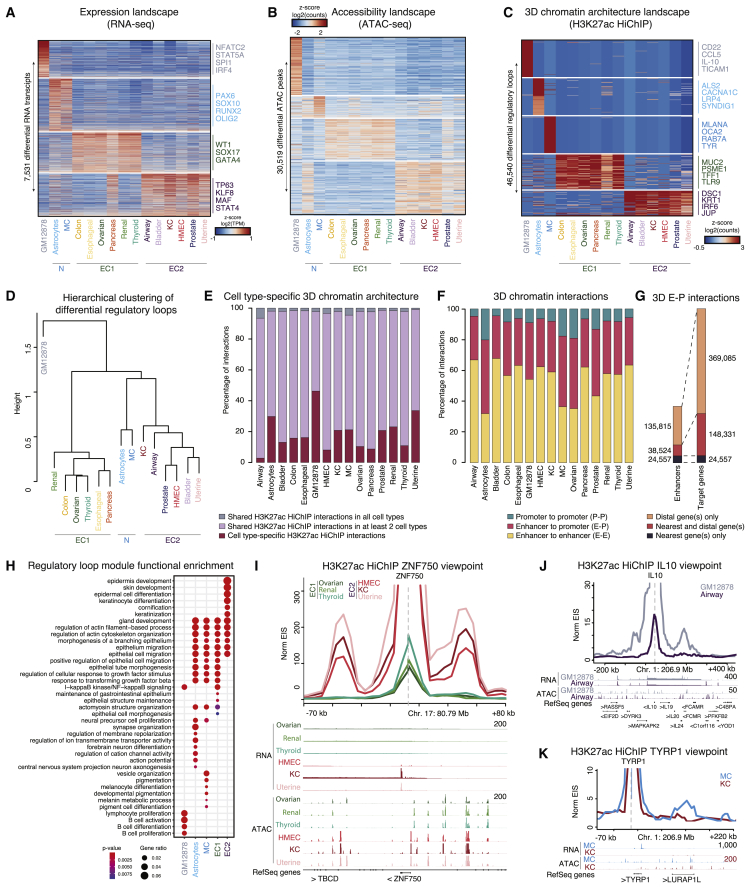
Figure 2.
Epigenomic landscape reveals distinct molecular subtypes of human cells
(A) RNA transcripts (rows) versus cell types (columns) of differential gene expression (log2 fold change >0.1, t test, FDR-adjusted p value <0.05).
(B) Heatmap of accessible peaks (rows) versus cell types (columns) indicating differential ATAC peaks. ATAC peaks with the highest inter-group SD shown.
(C) Heatmap of H3K27ac HiChIP loops (rows) versus cell types (columns) indicating differential loops. Differential loops with the highest inter-group SD shown.
(D) Hierarchical clustering of differential H3K27ac HiChIP loops.
(E) Bar plot depicting cell-type-specific 3D chromatin architecture and overlap between the 15 different cell types.
(F) Bar plot depicting distribution of P-P, E-P, and E-E interactions by cell type.
(G) Bar plot depicting putative enhancers and target genes identified in different E-P interaction types.
(H) Regulatory loop module functional enrichment using GO biological processes. EC1 and EC2 are grouped together. Dot color corresponds to the p value of the GO enrichment (hypergeometric test).
(I) Virtual 4C visualization at 5-kb resolution and RNA and ATAC-seq tracks centered at the ZNF750 TSS. > and < denote gene orientation on plus and minus DNA strand respectively.
(J) Virtual 4C visualization for IL10.
(K) Virtual 4C visualization for TYRP1. Related to Figures S1, S2, and Table S2.