Figure 3.
TF motif enrichment via footprinting cell-type CRMs
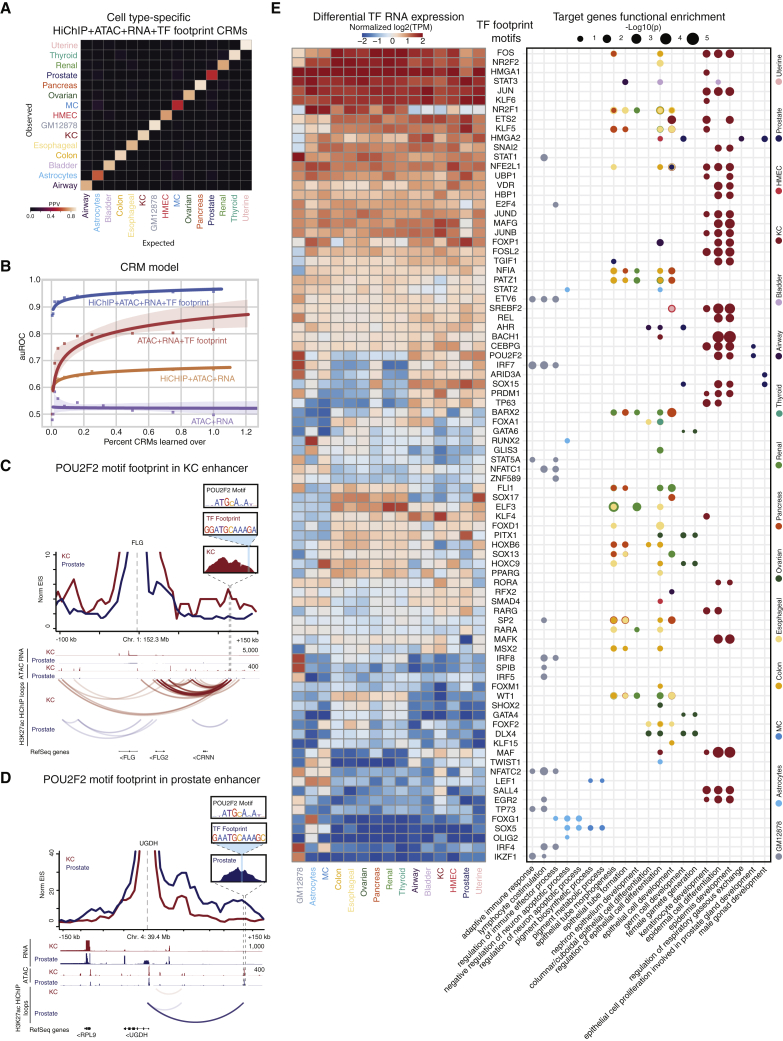
(A) Confusion matrix depicting the positive predictive value (PPV) for the cell type prediction model.
(B) Scatterplot showing auROC versus percentage of CRMs learned in the random-forest-based cell-type prediction model. Lines are fitted to the points using logistic regression.
(C) Virtual 4C visualization along with the POU2F2 position-weight matrix (PWM), TF footprint sequence, and surrounding ATAC peak centered at FLG.
(D) Virtual 4C visualization along with the POU2F2 PWM, TF footprint sequence, and surrounding ATAC peak centered at UGDH.
(E) Heatmap (left) depicts normalized log2(TPM) values for nominated TFs corresponding to motifs derived from TF footprinting analysis (rows) in the 15 cell types (columns). TFs are ordered by expression similarity. Dot plot (right) depicts GO enrichment for target genes (x axis) proximal or distally looped to TF footprint motifs in cell-type-specific CRMs (y axis). Dots are colored by cell type. Size corresponds to the −log10(p value) of the GO enrichment (hypergeometric test). Related to Figure S3.