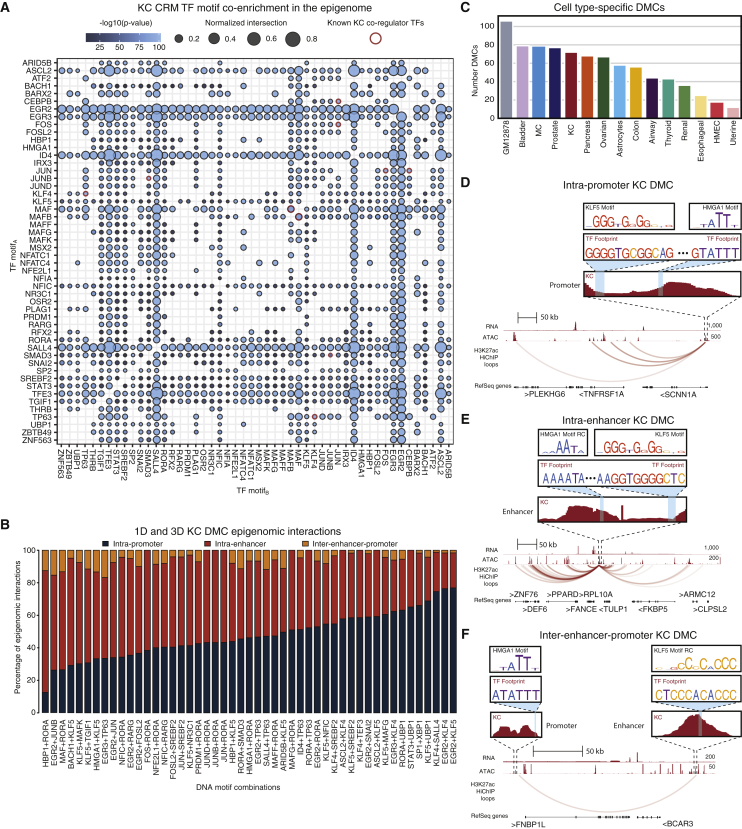
Figure 4.
Co-enrichment analysis reveals DMCs
(A) Co-enrichment dot plot of TF motifs within KC CRMs depicting putative cooperativity (Fisher’s exact, Bonferroni-corrected p < 0.05). Dots are colored by −log10(p value). Size corresponds to normalized number of shared genes. Red outlined dots indicate known cooperative KC TFs.
(B) Bar plot depicting the distribution of DMCs based on CRM epigenomic interactions for MPRA-tested KC DMCs.
(C) Bar plot of number of cell-type-specific DMCs in the 15 cell types.
(D) Genomic instance of intra-promoter KC DMC HMGA1+KLF5 at the SCNN1A TSS.
(E) Genomic instance of putative intra-enhancer KC DMC HMGA1+KLF5 looping to PPARD. RC, reverse complement.
(F) Genomic instance of putative inter-enhancer-promoter KC DMC HMGA1+KLF5 proximal to FNBP1L. Related to Figure S4 and Table S4.