Figure 6.
MPRAs identify regulatory DMCs in cancer
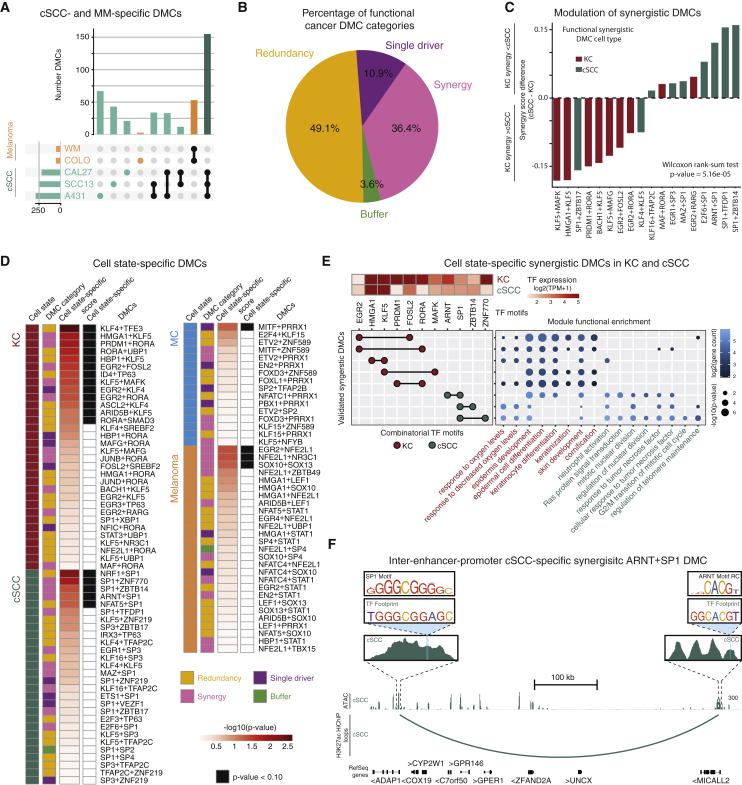
(A) Upset plot depicting number of DMCs determined from MM and cSCC cell lines and the size of their overlapping sets.
(B) Pie chart depicting percentage of functional DMC categories by MPRA in cSCC and MM cells.
(C) Bar plot showing the synergy score difference for KC- and cSCC-identified DMCs; p value based on a rank-sum Wilcoxon test.
(D) Left to right: panel colored by cell type/state; panel colored by functional DMC category; heatmap panel of −log10(p value) cell-type-/state-specificity score (STAR Methods); panel colored by cell-type-/state-specific expression (Wilcoxon rank-sum test p value <0.10).
(E) Top left: heatmap shows log2(TPM + 1) values for TFs in synergistic DMCs (columns) by normal KC- and cSCC-specific cell state (rows). Left: combinatorial TFs of the DMC (rows). Motifs that make up the DMC (columns) are circles with a black line connecting them. Circles are colored based on DMC cell state. Right: dot plot shows GO terms enriched for target genes (x axis) that utilize the DMC (y axis). Dots are colored by log2(target gene count). Dot sizes are the −log10(p value) of the GO enrichment. GO terms are colored by cell state biological processes.
(F) Genomic instance of putative inter-enhancer-promoter cSCC-specific synergistic DMC SP1+ARNT at ADAP1. Related to Figure S6 and Table S5.