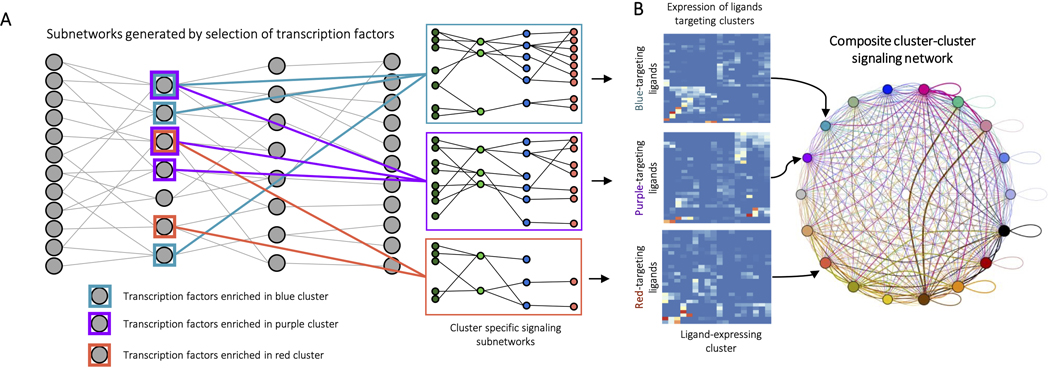
ED Fig. 6. Identification of intercluster signalling with Domino.
A, Identification of cluster-specific signaling subnetworks. Transcription factors enriched by cluster are identified by Wilcoxon rank sum and networks pruned for disconnected nodes to generate signaling subnetworks relevant for biological activation of clusters. B, Calculation of intercluster signaling networks. Once phenotypically relevant receptors are identified by cluster specific signaling subnetworks, cluster-cluster signaling scores are calculated by cluster averaged scaled expression of ligands present in cluster-specific subnetworks. Every potential cluster-cluster combination is scored, and these weights used to generate an intercluster signaling network.