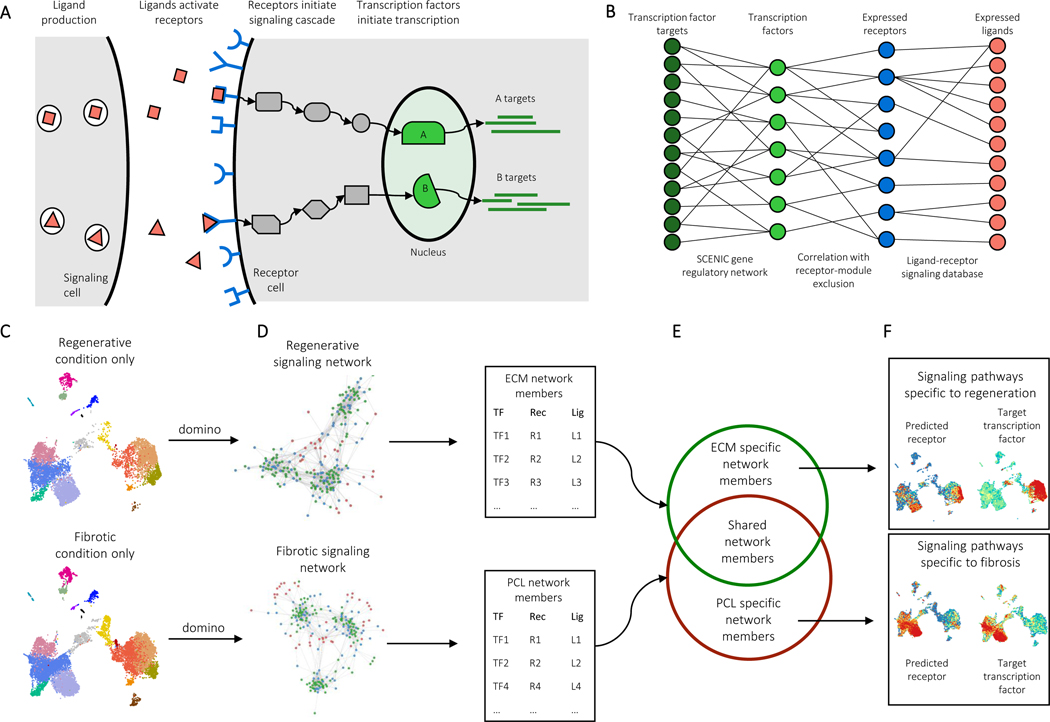
Figure 4: Generation of intercellular and intracellular signaling networks.
A, A model of biological ligand-receptor signaling. Ligands expressed in a signaling cell bind and activate receptors on a target cell. Subsequent protein-protein signaling triggers activation of transcription factors in the nucleus and expression of target genes. B, Reconstruction of a dataset-wide signaling network. SCENIC is used to estimate transcription factor gene regulatory modules, as well as transcription factor activation scores on a cell-by-cell level. Receptors expression levels are correlated with transcription factor activation scores across the entire dataset with exclusion of receptors present in the transcription factor modules. Public receptor-ligand databases and then queried to identify ligands capable of activation receptors. C, Identification of condition specific signaling patterns. In order to identify signaling specific to experimental conditions, the dataset is first split by condition, for example PCL and ECM treated cells. Domino is run on each to identify a signaling network specific to each condition. D, Members of signaling networks represent transcription factors, receptors, and ligands that are predicted as active in each condition. E, Comparison of network members from each condition identifies transcription factors, receptors, and ligands that are only predicted as active in one condition. F, Condition specific members can be used to identify signaling pathways which are only active or are differentially activated in an experimental condition.