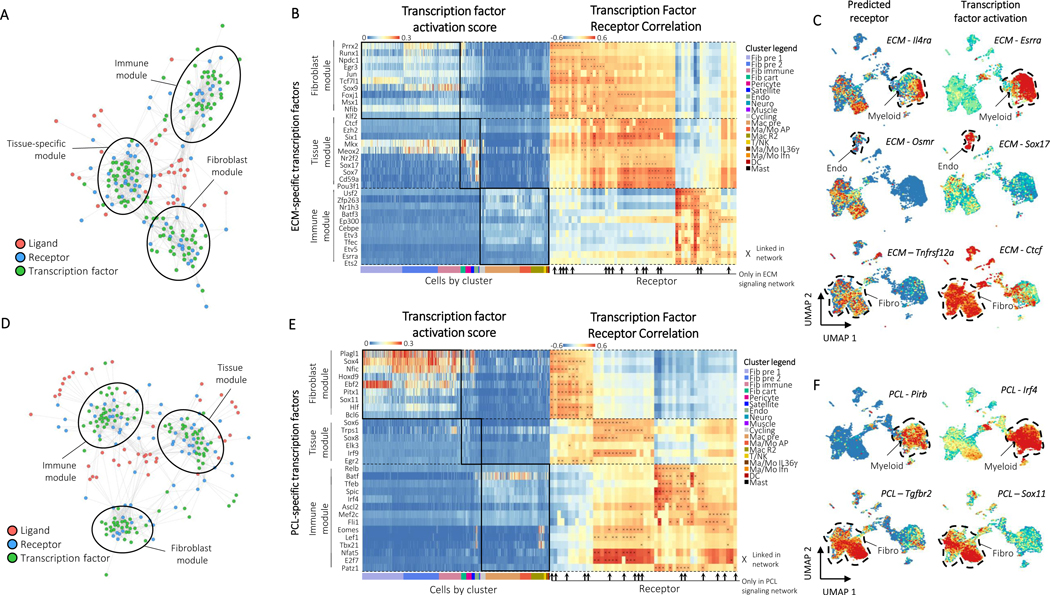
Figure 5: Domino identifies biomaterials condition specific signaling.
A, The ECM global signaling network. Three modules of receptors and transcription factors are readily apparent and labeled based on enrichment of transcription factors by cluster. B, Heatmaps of transcription factor activation score for ECM-specific transcription factors (left) and correlation of transcription factor activation score with receptor expression (right). Transcription factors are binned according to their membership to the fibroblast, tissue, or immune modules from the ECM global signaling network. Cells are ordered and colored according to their cluster. Receptors found only in the ECM condition are marked with arrows. Connections between receptor and transcription factors are marked with an ‘x’ on the correlation heatmap. C, Example feature plots of gene expression and activation scores for specific receptor-transcription factor pairs identified by domino in the ECM condition. D, The PCL global signaling network. Three modules of receptors and transcription factors are readily apparent and labeled based on enrichment of transcription factors by cluster. E, Heatmaps of transcription factor activation score for PCL-specific transcription factors (left) and correlation of transcription factor activation score with receptor expression (right). Transcription factors are binned according to their membership to the fibroblast, tissue, or immune modules from the PCL global signaling network. Cells are ordered and colored according to their cluster. Receptors found only in the PCL condition are marked with arrows. Connections between receptor and transcription factors are marked with an ‘x’ on the correlation heatmap. F, Example feature plots of gene expression and activation scores for specific receptor-transcription factor pairs identified by domino in the fibrotic (PCL) condition.