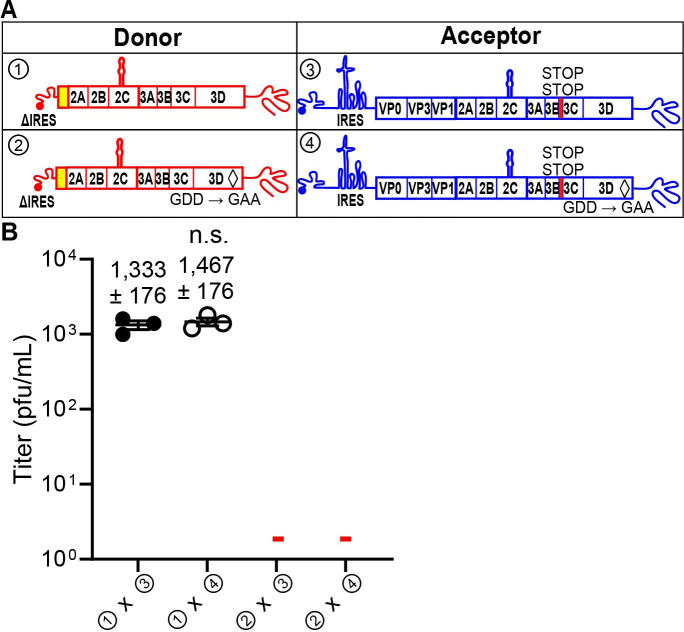
Fig 3. RNA virus recombinants are consistent with the donor RNA as the source of the PV RdRp, 3Dpol, and not the acceptor RNA.
(A) A mutation (GDD to GAA) producing an inactive PV RdRp, shown by a black diamond (◊), was introduced into either the donor or acceptor RNAs (② and ④). Insertion of two STOP codons (UAGUAA) after the 3B-coding sequence (3B STOP), indicated by a red rectangle, was introduced into the acceptor RNAs (③ and ④). (B) The indicated donor and acceptor RNAs were cotransfected into L929 cells. Yields of recombinant virus following transfection are shown (pfu/mL ± SEM; n = 3).—indicates that plaques were not detected (limit of detection: 2 pfu/mL). Statistical analyses were performed using unpaired, two-tailed t test (n.s. indicates not significant). Viral recombinants were recovered only when the donor had an intact 3D gene encoding an active RdRp (① x ③, ① x ④); viral recombinants were not recovered when the donor RNA encoded an inactive RdRp (② x ③, ② x ④). There was no impact on viral recombinants produced when the acceptor RNA had stop codons upstream of the 3D gene encoding an active or inactive RdRp (① x ③, ① x ④). Numerical data provided as Supporting information (S1 Data). PV, poliovirus; RdRp, RNA-dependent RNA polymerase.