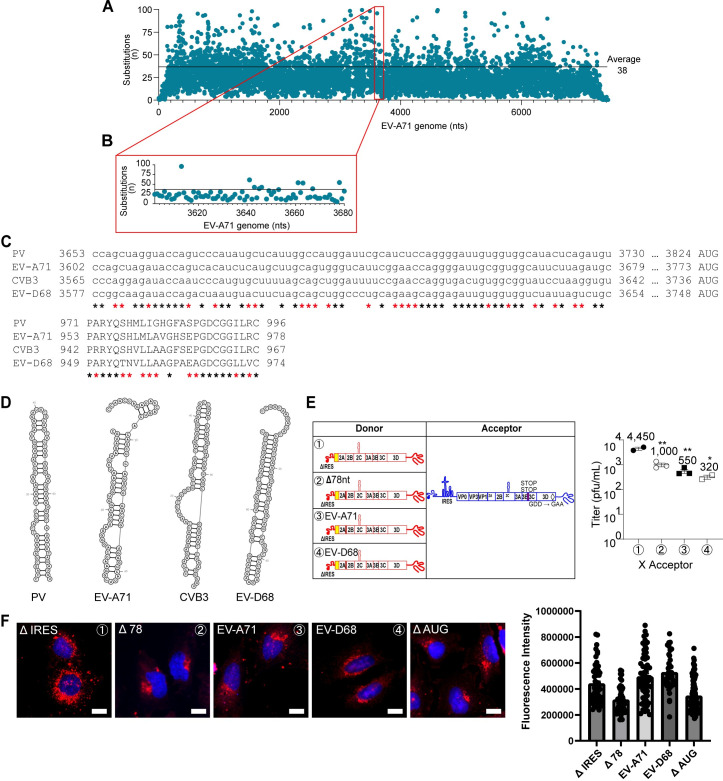
Fig 11. Identification of an RNA sequence upstream of the AUG codon required for eIF2A/2D-dependent translation.
(A) Nucleotide substitutions at each position across the EV-A71 genome have been defined by deep RNA sequencing. The average substitution frequency is 38. Some regions exhibit a lower average; these regions of the genome may encode cis-acting RNA elements. Numerical data provided as Supporting information (S1 Data). The entire data set is available at the GEO repository under accession number GSE183959. (B) The region from 3,602–3,679 of the EV-A71 genome exhibits a below-average frequency of nucleotide substitutions. This region corresponds to a sequence 78-nt upstream of the AUG used for eIF2A/2D-dependent translation. Numerical data provided as Supporting information (S1 Data). (C) Alignment of the corresponding nucleotide sequence and amino acid sequence reveals moderate to high sequence identity among enterovirus (PV, EV-A71, CVB3, and EV-D68). Black asterisks indicate conservation across all enteroviruses; red asterisks indicate conservation in three of the four enteroviruses. (D) RNA secondary structure is predicted in this region for all enteroviruses. The RNAfold algorithm was used. The details of the fold varied across the enteroviruses more substantially than sequence might predict. (E) Comparison of infectious PV produced using donor RNAs harboring a deletion of the 78-nt sequence (Δ78 nt) or containing the corresponding sequences from EV-A71 or EV-D68. The acceptor RNA used does not support translation of 3CD or an active polymerase. Virus produced (pfu/mL ± SEM; n = 3) from each cotransfection is shown. Statistical analyses were performed using unpaired, two-tailed t test (* indicates p < 0.05, ** indicates p < 0.01; n.s. indicates nonsignificant). The sequence contributes to production of virus. Numerical data provided as Supporting information (S1 Data). (F) Contribution of the 78-nt sequence to IRES-independent translation indirectly by monitoring PI4P levels and localization as described in the legend to Fig 7. Donor RNAs of panel E were used, in addition to the ΔAUG donor RNA used in Fig 9 that has a strong defect to virus production. Quantitation of PI4P staining in approximately 60 cells selected randomly from three separate fields expressed as fluorescence intensity is shown. A significant reduction of PI4P/translation of the genome (p < 0.0001) was observed for Δ78 and ΔAUG only. Scale bars are equivalent to 10 μm. Numerical data provided as Supporting information (S1 Data). GEO, Gene Expression Omnibus; IRES, internal ribosome entry site; PI4P, phosphatidylinositol-4-phosphate; PV, poliovirus.