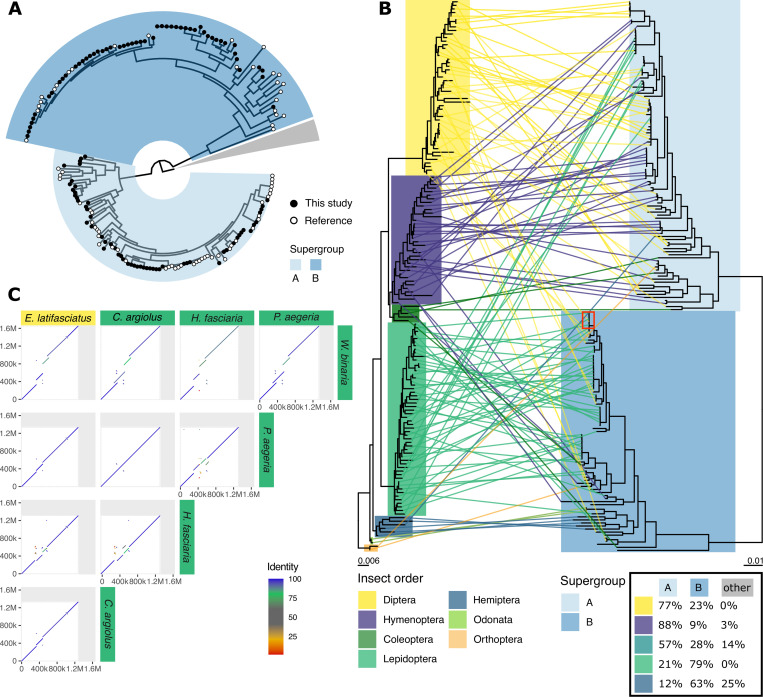
Fig 2. Wolbachia DToL genomes expand known phylogeny.
(A) Circular phylogeny of supergroup A and B Wolbachia, visualised with the root placed between the A and B supergroups and the remaining supergroups (C, D, E, F, J, S; nodes collapsed as grey wedge), highlighting newly sequenced genomes (black tip labels) and genomes from public databases (white). (B) Incongruence between host topology (left) and supergroup A and B Wolbachia topology (right) is shown as a tanglegram. Overview of the supergroups infecting diverse insect orders is given in a table (inset, bottom right). A red box is drawn to point to a host switching event; see panel C. (C) Example of a host switching event, where the Wolbachia of the hoverfly Eupeodes latifasciatus has high nuclear sequence identity and genome colinearity to four Wolbachia genomes assembled from Lepidoptera.