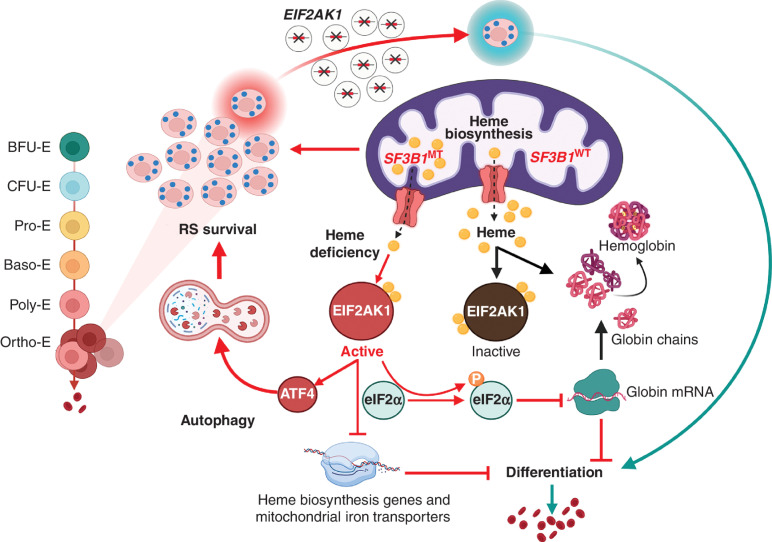
Figure 5.
Proposed model. Under normal physiologic conditions, heme binds to EIF2AK1 and represses its activation. In conditions that limit heme production, such as those induced by SF3B1MT in terminally differentiated erythroblasts, EIF2AK1 is activated. EIF2AK1 pathway activation promotes RS survival and inhibits erythroid maturation by increasing the expression of ATF4, which in turn upregulates the expression of genes involved in autophagy, transcriptionally inhibits genes involved in heme biosynthesis and mitochondrial iron transport, and translationally inhibits globin production. Targeting EIF2AK1 pathway activation by depleting EIF2AK1 rescues erythroid differentiation and red blood cell production.