Amplified EPOR/JAK2 with multi-hit TP53 lesions define a unique subtype of AEL showing extreme erythroid differentiation with activated STAT5 and aggressive clinical courses, for which a potential therapeutic role of JAK2 inhibition is suggested.
Abstract
Acute erythroid leukemia (AEL) is a unique subtype of acute myeloid leukemia characterized by prominent erythroid proliferation whose molecular basis is poorly understood. To elucidate the underlying mechanism of erythroid proliferation, we analyzed 121 AEL using whole-genome, whole-exome, and/or targeted-capture sequencing, together with transcriptome analysis of 21 AEL samples. Combining publicly available sequencing data, we found a high frequency of gains and amplifications involving EPOR/JAK2 in TP53-mutated cases, particularly those having >80% erythroblasts designated as pure erythroid leukemia (10/13). These cases were frequently accompanied by gains and amplifications of ERG/ETS2 and associated with a very poor prognosis, even compared with other TP53-mutated AEL. In addition to activation of the STAT5 pathway, a common feature across all AEL cases, these AEL cases exhibited enhanced cell proliferation and heme metabolism and often showed high sensitivity to ruxolitinib in vitro and in xenograft models, highlighting a potential role of JAK2 inhibition in therapeutics of AEL.
Significance:
This study reveals the major role of gains, amplifications, and mutations of EPOR and JAK2 in the pathogenesis of pure erythroleukemia. Their frequent response to ruxolitinib in patient-derived xenograft and cell culture models highlights a possible therapeutic role of JAK2 inhibition for erythroleukemia with EPOR/JAK2-involving lesions.
This article is highlighted in the In This Issue feature, p. 369
INTRODUCTION
Attracting continuous attention of generations of hematologists due to its unique morphologic feature of conspicuous erythroid proliferation (1–6), acute erythroid leukemia (AEL) represents a rare subtype of acute myeloid leukemia (AML), accounting for 0.5% to 1.5% of AML cases (7, 8). Since it was first described (9, 10), the definition of AEL has undergone changes over time with frequent confusions with other AML categories and myelodysplastic syndromes (MDS). According to the previous classification system of the World Health Organization (WHO; WHO 2001), AEL included two major categories on the basis of their morphologic features: those having pure erythroid components (>80 of erythroblasts; pure erythroid leukemia, PEL) and those with more myeloid components (≥50% and <80% erythroblasts and ≥20% myeloblasts in nonerythroid cells; erythroid/myeloid leukemia, EML; refs. 2, 4). However, in the most recent revision of the WHO classification (11), the diagnosis of AEL has been revised to include only PEL, while excluding EML, with the latter now classified as other forms of either of AML or MDS, depending on the percentages of myeloblasts. Despite many historical changes and confusion regarding the definition of AEL, which often included other subtypes of AML, such as AML with myelodysplasia-related changes (AML-MRC) and AML not otherwise specified (AML-NOS; ref. 12), previous genetic studies have consistently demonstrated frequent mutations in TP53, NPM1, STAG2, transcription factors, and chromatin modifiers (13–18). However, also commonly mutated in non-AEL cases (19), these mutations may not necessarily explain the unique erythroid-biased phenotype of AEL or the distinction between AEL and nonerythroid AML (non-AEL). For example, TP53 mutations, particularly multihit mutations in combination with extensive aneuploidy (20), are found in a wide variety of myeloid neoplasms, including AEL and other AML, MDS, and MDS/MPN, and are uniformly associated with a dismal prognosis, regardless of diagnosis (16, 18, 21). Moreover, the lack of promising druggable targets prevents improvement of the clinical outcome of AEL, although a potential role of hypomethylating agents and other compounds has been discussed for TP53-mutated cases (22–24).
To clarify the mechanism of erythroid predominance in AEL and also identify molecular targets for the development of novel therapeutics for AEL, we enrolled a total of 124 AEL patients as per the WHO 2001 criteria and characterized their somatic mutations, copy-number alterations (CNA), structural variations (SV), and/or gene-expression profiles, which were compared with those in 409 cases non-AEL (WHO 2001) and 229 with MDS with excess blasts (MDS-EB; WHO 2017) cases without erythroid hyperplasia (see Methods section). We identified frequent focal gains and/or amplifications of genes implicated in erythroid proliferation and differentiation, particularly EPOR and JAK2, which resulted in enhanced STAT5 signaling and promoted cell proliferation. Finally, we demonstrated a potential therapeutic role of JAK2 inhibition, using in vitro culture of AEL cell lines and in vivo AEL-derived patient-derived xenograft (PDX) models.
RESULTS
Unbiased Sequencing Analysis of AEL
To identify previously unknown genetic lesions in AEL, we first performed whole-genome sequencing (WGS; n = 20) and/or whole-exome sequencing (WES; n = 27) in a subset (n = 35) of our AEL cases, including 6 PEL, 16 EML, and 13 other AEL cases. We identified a median of 0.569 and 0.504 SNVs/Mb/sample and 6.75 × 10−3 and 0 indels/Mb/sample in PEL and EML cases, respectively, which were largely comparable with the values previously reported for the TCGA AML cases (7). In total, 58 genes were recurrently mutated in the coding regions, of which TP53 (n = 15), STAG2 (n = 6), KMT2A (n = 6), TET2 (n = 5), NPM1 (n = 5), and WT1 (n = 4) were most frequently affected (Supplementary Fig. S1A and S1B). A median of 20.5 SVs were detected in WGS, including large deletions, tandem duplications, and inversions. However, the number of SVs substantially differed between samples, ranging from 5 to 399 depending on the sample, where TP53-mutated samples had significantly higher numbers of SVs (median: 24/sample, range, 11–399) than TP53-intact samples (median: 15/sample, range, 5–26; P = 1.67 × 10−2; Supplementary Fig. S1C). TP53-mutated samples also had significantly higher numbers of CNAs (median: 10.5/sample; range, 6–23) than unmutated samples (a median of 0/sample, range, 0–1) in copy-number (CN) analysis based on WGS/WES (P = 1.30 × 10−7). Of particular interest among these SVs and CNAs were focal gains/amplifications recurrently affecting the JAK2 (9p21), EPOR (19p13), and/or ERG/ETS2 (21q22) loci (n = 5), because these genes are implicated in the regulation of erythroid differentiation/proliferation. Multiple loci were affected in seven cases (Supplementary Fig. S1B). In three cases, amplification of EPOR and ERG/ETS2 was associated with chromothripsis events involving both loci, as reported previously (ref. 25; Fig. 1A and B; Supplementary Fig. S2).
Figure 1.
Gain/amplification affecting chromosomes 9p, 19p, and 21q in AEL. A, Complex SVs affecting chromosomes 19 and 21 for a representative case (UPN091). Left, CNVs and SVs are shown along chromosome ideograms. Chromosomes 19 (Chr 19) and 21 (Chr 21) are highlighted. Chromothripsis lesions involving chromosomes 19 and 21 are shown on the right. Yellow rectangles indicate the loci of EPOR and ERG/ETS2. B, An example of CNV (UPN093). The top and bottom panels show typical CN amplifications in chromosomes 9p, 19p, and 21q identified by WGS and the ideogram of the corresponding chromosomes.
Genomic Landscape of AEL
To confirm these recurrent SNVs and SVs/CNAs detected in WGS/WES, we analyzed diagnostic samples from all 121 AEL cases, together with 214 non-AEL cases, using targeted−capture sequencing with a mean depth of 557× and 615×, respectively (Supplementary Figs. S1 and S3A; Supplementary Table S1). Diagnosis of the 121 adult AEL cases was made according to the 2001 WHO classification, which included 13 PEL and 82 EML cases, of which 3 turned out to be therapy-related (26). Due to the lack of detailed information, the subcategories (PEL or EML) were not specified in the remaining 26 cases (Supplementary Table S2). The target gene panel included a high-density bait set designed to sensitively capture focal gains/amplifications of EPOR, JAK2, and ERG/ETS2 loci, in addition to common mutations in myeloid neoplasms (refs. 7, 8, 19, 27–29; Supplementary Table S3). We also designed a number of baits to capture 1,216 SNP sites to enable sequencing−based genome-wide CN analysis (20). Combining an additional 3 AEL cases from the TCGA AML data set, the initial results were largely recapitulated, where mutations most frequently affected TP53 (40.3%), followed by STAG2 (20.1%), KMT2A (20.2%), TET2 (16.9%), and NPM1 (14.5%; Fig. 2; Supplementary Fig. S3B and S3C; Supplementary Table S4). Also including an additional 195 non-AEL cases from TCGA, mutational profiles were shown to be substantially different between AEL (n = 124) and non-AEL (n = 409) cases (Supplementary Table S1); TP53 and STAG2 mutations and KMT2A-PTD were overrepresented in AEL, whereas those affecting FLT3, NRAS, and DNMT3A were significantly underrepresented in AEL (Supplementary Fig. S3D; Supplementary Table S5). Conspicuously, accounting for 71.8% of all AEL cases, TP53, NPM1, and STAG2 mutations were almost mutually exclusive (Supplementary Fig. S4A), and STAG2 mutations showed a strong association with KMT2A-PTD and CEBPA mutations. Based on these mutually exclusive and cooccurring relationships, AEL cases were clustered into four genetically discrete groups, groups A−D (Fig. 2).
Figure 2.
Global genetic profile in acute erythroleukemia. Genomic landscape of 124 AEL cases. Each column indicates a patient. The heatmap demonstrates mutations, SVs, and CNAs. Genetic groups were clustered according to the status of genetic events. Cases with AEL were divided into four groups based on the correlation of frequent mutations and SVs [A:TP53-mutated AEL, B:NPM1-mutated AEL, C:STAG2-mutated but NPM1-intact AEL, D: other (triple-negative, TN) AEL]. UPD: uniparental disomy.
Genetic Feature of AEL Subgroups
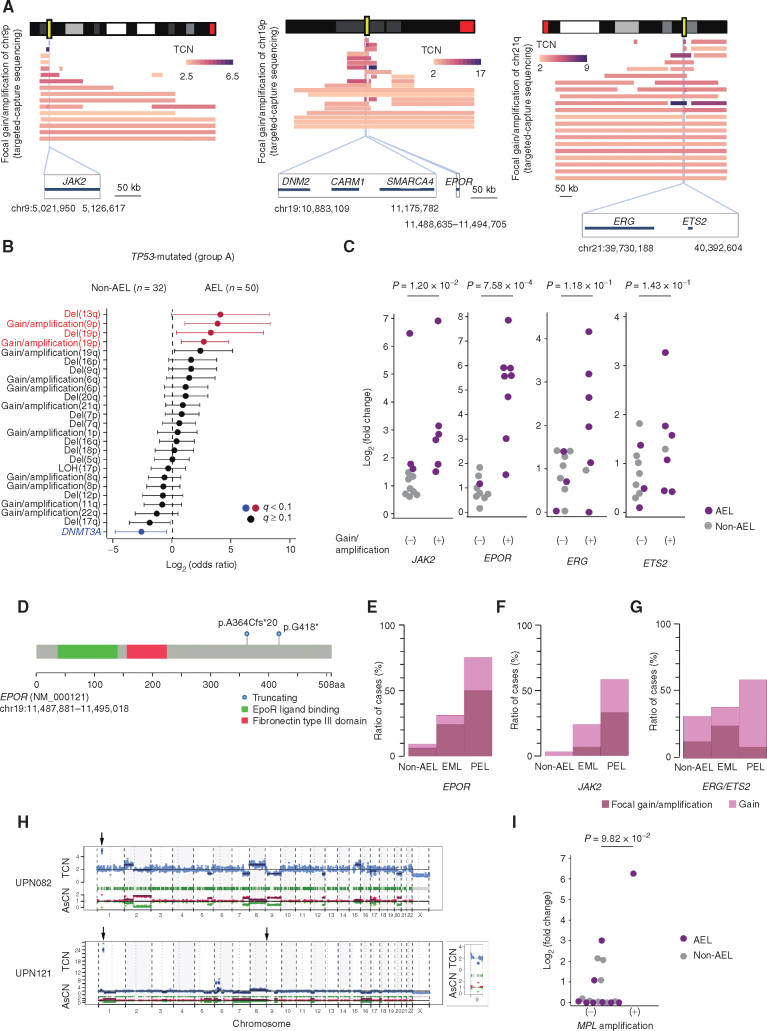
Accounting for 40.3% (n = 50) of the entire AEL cohort, group A is characterized by TP53 mutations. No germline TP53 mutations were detected in group A cases for which germline DNA was obtained (23 of 50). All group A cases accompanied complex karyotype (CK) and/or extensive CNAs (≥3), suggestive of CK (“CK-like”), which were rarely found in TP53-unmuated AEL cases. In line with a study in TP53-mutated MDS (20), most cases (n = 46) had multihit TP53 lesions, including multiple mutations (n = 14) and/or mutation(s) plus 17p loss of heterozygosity (LOH; n = 33). TP53 mutations were also found in non-AEL AML but were much less common [32/409 (7.8%) in our cohort and 6.2% in a larger series; ref. 19]. Although the CN profile in TP53-mutated cases was largely similar between AEL and non-AEL with frequent involvement of −5/del(5q) (69.3% vs. 68.8%), 17p LOH (67.3% vs. 71.9%), −7/del(7q) (66.7% vs. 56.3%), +21q (42.8% vs. 28.1%), +8q (30.6% vs. 40.6%), and del(16q) (30.6% vs. 25%), several lesions, such as +19p (40.8% vs. 9.38%), +9p (32.7% vs. 3.13%), +19q (26.5% vs. 6.25%), and del(13q) (20.4% vs. 0%), were more frequently found in, and therefore characteristic of, TP53-mutated AEL cases (Fig. 3A and B; Supplementary Fig. S4B; Supplementary Table S6). Among these, however, gains/focal amplifications involving the EPOR (19p13), JAK2 (9p21), and ERG/ETS2 (21q22) loci were almost exclusively found in group A, in which ∼60% (29/50) of the patients harbored one or more of these lesions, whereas they were rarely found in other AEL cases (1/74; group B−D; Figs. 2 and 3A; Supplementary Fig. S5). In particular, those affecting EPOR and JAK2 were highly specific to TP53-mutated AEL and rarely found in non-AEL, even in TP53-mutated cases (25/50 vs. 4/32; P = 7.49 × 10−4). In qPCR analysis, these gains/focal amplifications of EPOR and JAK2 were significantly associated with overexpression of the affected genes (Fig. 3C). Moreover, we identified activating mutations in JAK2 (p.V617F) and EPOR (p.A364fs and p.G418*; refs. 30, 31) in three cases with TP53-mutated AEL, including two cases (UPN093 and UPN094) with concomitant focal amplification/gain and activating EPOR mutations (Fig. 3D; Supplementary Fig. S5). Combined, these findings support that EPOR and JAK2 are the functional targets of gains/focal amplifications involving 19p and 9p, respectively. Of note, both genes were more frequently affected by gains/focal amplifications in PEL with TP53 mutations than EML with TP53 mutations [EPOR: 10/12 (83%) vs. 9/29 (31%), P = 1.6 × 10−2, and JAK2: 7/12 (67%) vs. 7/29 (24%), P = 6.8 × 10−2; Fig. 3E and F], suggesting their link to the PEL phenotype. Supporting this are three cases in which transformation from MDS to AEL was accompanied by increasing copy numbers of the EPOR locus (UPN089 and UPN105) or disappearance of 9p gain coincided with loss of AEL phenotype (UPN086; Supplementary Fig. S6A–S6C; Supplementary Table S7). By contrast, gains/focal amplifications of ERG/ETS2 were less specific to AEL and also found in non-AEL cases at a comparable frequency (Fig. 3G). Finally, we noted two cases with amplification and overexpression of MPL (Fig. 3H and I), also underscoring a role of upregulated JAK2/STAT signaling in AEL pathogenesis.
Figure 3.
AEL-specific genomic lesions in TP53-mutated erythroleukemia. A, Frequent gains/amplifications in chromosomes 9p, 19p, and 21q. CNAs were identified by targeted-capture sequencing/WES. The top and bottom panels show the ideogram of the corresponding chromosomes and the range of affected regions based on targeted-capture sequencing, respectively. Vertical light blue lines and yellow rectangles indicate commonly amplified regions and affected genes, respectively. The color gradient represents the TCN normalized by the TCF. Genes located in commonly amplified regions are shown in the lower part of each ideogram. B, Forest plot showing the differences of mutational profiles between AEL (n = 50) and non-AEL (n = 32) with TP53 mutation. Colors indicate affected lesions with >95% specificity for AEL (red) or >95% specificity for non-AEL (blue). Odds ratio and q-values are shown by the mark and color of forest plots. C, Association between DNA amplification and upregulation of expression. The results of RT-qPCR of AML samples with or without gains/amplifications of the indicated genes were compared. AEL and non-AEL are shown as purple and gray dots, respectively. Hollow dots indicate the cases with simple gain. GAPDH was used as an internal control. D,EPOR-truncating mutations detected in the AEL cohort are indicated. E–G, Differential prevalence of frequent genetic events in TP53-mutated cases. The bar plot demonstrates frequencies of gains/amplifications in EPOR (E), JAK2 (F), and ERG/ETS2 (G) between PEL, EML, and non-AEL indicated by the red (focal gain/amplification) and pink (gain) box. H, CNAs detected by the CNACS algorithm of AEL with MPL amplifications. The horizontal axis indicates each chromosome. The vertical axis indicates the TCN (top) and the allele-specific copy number (AsCN; bottom). Arrows indicate focal gains/amplifications, including JAK2 (chr 9) or MPL (chr 1). The magnified image of chromosome 9 of UPN121 is shown on the right. I, Association between DNA amplification and upregulation of expression. The results of RT-qPCR of AML samples with or without amplifications of MPL were compared. AEL and non-AEL are shown as purple and gray dots, respectively. GAPDH was used as an internal control.
Group B (n = 18, 14.5%) was defined by the presence of NPM1 mutations (Fig. 2), including a case with NPM1–MLF1 fusion associated with t(3;5)(q25;q34). Although NPM1 mutations also define a unique molecular subtype among non-AEL cases (20%–30%; refs. 7, 8, 19), the profile of accompanying mutations substantially differed between AEL and non-AEL cases. Although representing the most frequent mutations accompanying NPM1-mutated non-AEL (55.4%; refs. 7, 8, 19), FLT3 mutations were much less common (16.7%) in NPM1-mutated AEL. Instead, PTPN11 mutations, which are less common in NPM1-mutated non-AEL, were found in 50% of NPM1-mutated AEL cases (Supplementary Fig. S7A and S7B; Supplementary Table S8). Of note, 9 of 11 PTPN11 mutations in AEL were identified in NPM1-mutated cases (Fig. 2). These findings suggest that these AEL-specific co-occurring mutations might have a significant impact on erythroid predominance in NPM1-mutated AEL, particularly, in EML.
Characterized by mutated STAG2, group C accounted for 17.7% of all AEL cases (Fig. 2), the majority (n = 17/22) of which had KMT2A-PTD. Conversely, 17 of 23 cases with KMT2A-PTD accompanied STAG2 mutations. Rarely found in STAG2-mutant non-AEL (1/16), KMT2A-PTD in combination with mutated STAG2 might be responsible for the AEL phenotype (Supplementary Fig. S7C and S7D; Supplementary Table S9). Other common mutations in group C cases included TET2 (31.8%), CEBPA (31.8%), and SRSF2 (22.7%; Supplementary Fig. S7C), of which CEBPA mutations were highly enriched in this AEL subtype (7/22 vs. 1/102; Fig. 2).
None of the remaining 34 (27.4%) group D cases harbored mutations in TP53, STAG2, or NPM1. Most frequently mutated in these “triple-negative (TN)” cases were genes for chromatin/histone modification (50.0%), including ASXL1, BCOR, KMT2A/2C, DNA methylation (47.1%), RNA splicing (32.3%), and RUNX1 and other transcription factors, and therefore correspond to chromatin/spliceosome-mutated AML (ref. 19; Supplementary Fig. S7E). Compared with similar TN-non-AEL, group D was enriched for mutations in BCOR, USP9X, PHF6, U2AF1, ASXL1, RUNX1, and TET2, of which USP9X and BCOR mutations showed the strongest enrichment; USP9X mutations were observed in 5 of 34 cases with TN-AEL but found in 5 of 266 TN-non-AEL cases (q = 5.48 × 10−3; Supplementary Fig. S7F; Supplementary Table S10). In most cases, mutations were truncating (3 frameshift, 1 nonsense, 1 splice site, and 1 missense; Supplementary Fig. S7G), suggesting that loss of its function might be implicated in the pathogenesis of TN-AEL.
To summarize, AEL comprises 4 categories that correspond to non-AEL counterparts characterized by mutated TP53 with aneuploidy, NPM1, and chromatin/spliceosome-mutated with or without STAG2 mutations. Each category has AEL-specific comutations/abnormalities, such as focal gains/amplification of EPOR/JAK2 with or without ERG/ETS2 lesions, PTPN11 mutations, KMT2A-PTD, and USP9X and BCOR mutations, respectively. It should also be noted that many of our EML cases (61/82) had <20% total blasts and therefore classified as MDS according to the most updated WHO classification (11). In line with this, these cases showed a significant enrichment of mutations in the genes that were more frequently seen in MDS or secondary AML (sAML) than primary AML, such as those affecting TP53, STAG2, BCOR, ASXL1, and splicing factors (32, 33). However, despite the similarity to MDS and MDS-derived sAML, these cases with <20% total blasts had distinct mutational profiles compared with other MDS cases. Overall, EML cases with <20% total blasts are enriched for mutations in CEBPA, IDH1, WT1, and STAG2, and KMT2A-PTD compared with 229 MDS-EB cases without erythroid hyperplasia (Supplementary Fig. S8A; Supplementary Table S11). Moreover, TP53-, STAG2-, and NPM1-mutated EML cases with <20% total blasts were enriched for gains/amplifications of EPOR, JAK2, and other chromosomal lesions, KMT2A-PTD, and CEBPA mutations, and PTPN11 mutations, compared with respective MDS-EB cases without erythroid hyperplasia (Supplementary Fig. S8B–S8D; Supplementary Tables S12–S14). Also, a subset of patients with group D AEL with <20% total blasts had higher frequencies of mutations in USP9X, WT1, SF3B1, BCOR, and RUNX1 (Supplementary Fig. S8E; Supplementary Table S15). Thus, these AEL cases <20% of total blasts are considered genetically distinct from other MDS-EB cases without erythroid hyperplasia.
Gene-Expression Profile
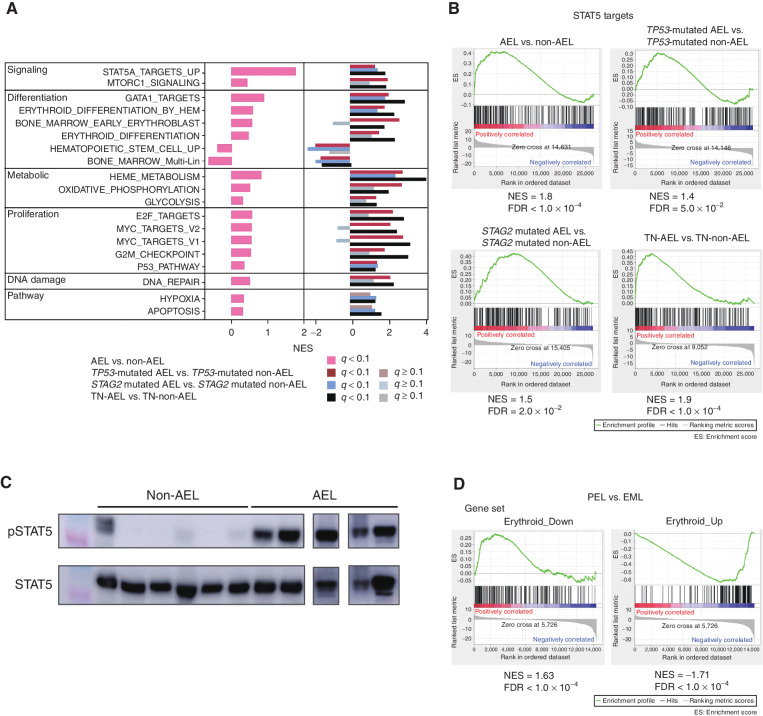
To understand AEL pathogenesis in terms of gene expression, we analyzed transcriptome data of whole BM cells or PDX cells from 23 AEL samples (n = 21 from the in-house cohort and n = 2 from TCGA cohort), which were compared with those from 213 non-AEL cases (Supplementary Table S1). As a whole, AEL showed a prominent upregulation of STAT5A target genes compared with non-AEL (Fig. 4A and B). The enrichment of STAT5 target genes in AEL compared with non-AEL was significant, even when the comparison was made within individual subcategories of AEL and corresponding non-AEL, i.e., TP53-mutated, NPM1-mutated, STAG2-mutated, and other AEL and non-AEL cases (34, 35). The activated STAT5 in AEL was further confirmed by an enhanced phosphorylation of STAT5 in AEL-derived PDX cells in western blot analysis, compared with non–AEL-derived PDX cells (Fig. 4C). Other features of the AEL expression profile included an enhanced expression of gene sets implicated in mTORC1 signaling, erythroid differentiation, GATA1 target genes, heme metabolism, cell proliferation, and DNA repair and downregulation of genes related to hematopoietic stem cells and multilineage progenitors (Fig. 4A). These features, including enhanced STAT5 signaling, were also observed when the comparison was made between individual AEL subtypes (group A, C, and D) and their non-AEL counterparts (Fig. 4A). They were more pronounced in the cases with gains/amplifications of EPOR/JAK2/ERG/ETS2 (n = 9), compared with those without gains/amplifications of EPOR/JAK2/ERG/ETS2 (n = 4), even though they were still observed in the latter cases in comparison with TP53-mutated non-AEL cases.
Figure 4.
Difference in RNA expression between AEL and non-AEL and between PEL and EML. A, Gene sets enrichment analysis of AEL (n = 23) vs. non-AEL (n = 213). Normalized enrichment scores (NES) between AEL and non-AEL using hallmark gene sets, gene sets involved in STAT5 targets, and erythroid differentiation with false discovery rate (FDR) q-value < 0.10. The comparison between AEL and non-AEL with TP53 mutation and STAG2 mutation, and without TP53, NPM1, and STAG2 mutation are also shown. B, The results of gene set enrichment analysis (GSEA) using a gene set of STAT5 targets are shown. C, Representative western blot results (experiments were performed in triplicate). Immunoblot shows the phosphorylation status of STAT5 in six non-AEL controls and five AEL with TP53 mutations. D, GSEA shows that compared with that of EML, the expression PEL is positively correlated with the gene set of Erythroid_Down (left) and negatively correlated with the gene set of Erythroid_UP (right).
Next, to understand the phenotypic difference between PEL and EML on the basis of gene expression, we compared gene-expression profiles between PEL and EML. Because a prominent proliferation of immature erythroblasts is a cardinal feature of PEL compared with EML, we first constructed two gene sets, which are most upregulated (Erythroid_Up; n = 200) and downregulated (Erythroid_Down; n = 200) during normal erythroid differentiation, respectively, according to a published gene-expression analysis of different stages of erythroblasts (ref. 36; Supplementary Table S16). Then, we evaluated the enrichment of each gene set in the differentially expressed genes between PEL (n = 5) and EML (n = 16) samples. In agreement with the prominent maturation arrest in PEL, we observed a significant enrichment of the Erythroid_Up and Erythroid_Down gene sets in significantly upregulated and downregulated genes in PEL, respectively (Fig. 4D).
Prognostic Impacts of Common Genetic Lesions
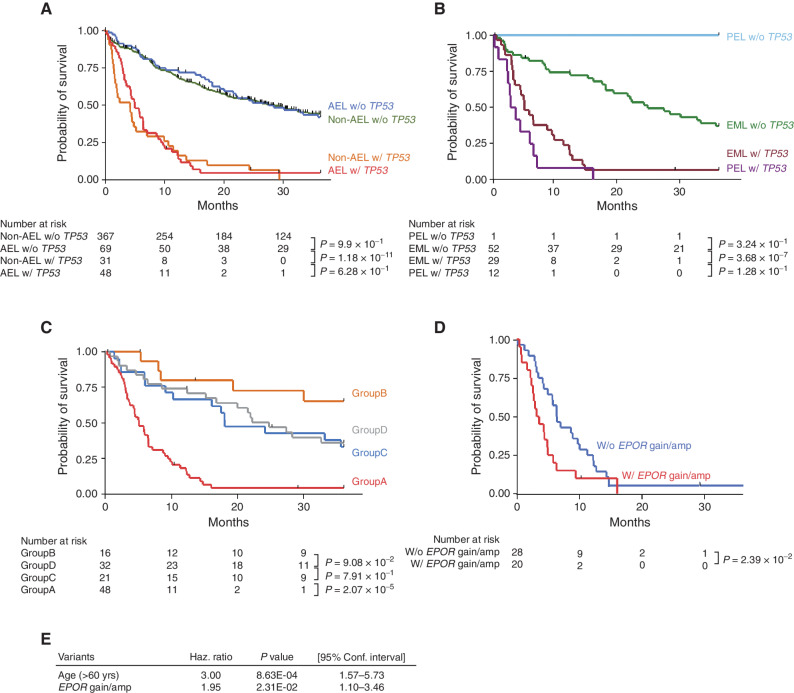
As a whole, AEL cases exhibited a substantially shorter overall survival (OS) compared with non-AEL cases (Supplementary Fig. S9A). However, this apparent difference in OS is largely explained by a higher representation of TP53-mutated cases in AEL (40.3 vs. 7.82%, respectively; Supplementary Fig. S3D); when the cohort was stratified by TP53 mutation status, no significant difference in OS was observed between AEL and non-AEL cases (Fig. 5A). Similarly, a significantly poorer prognosis of PEL than EML (P = 1.44 × 10−3) is also explained by a significant enrichment of TP53-mutated cases in PEL (Fig. 5B; Supplementary Fig. S9B). Of note, all but one PEL case were classified into group A, suggesting that PEL is essentially exclusively a TP53-mutated disease with a dismal prognosis. Excluding the TP53-mutated subtype, other AEL subtypes (groups B–D) showed similar OS (Fig. 5C). No significant difference in OS was observed among AEL subtypes and their non-AEL counterparts, TP53-, STAG2-, and NPM1-mutated, and TN subtypes (Supplementary Fig. S9C–S9E). Multivariable analysis of OS in the group A patients (see Methods for detail) revealed that age [>60 years; hazard ratio (HR): 3.00; 95% CI, 1.57−5.73, P = 8.63 × 10−4) and gains and/or focal amplifications of the EPOR locus on 19p (HR: 1.95; 95% CI, 1.10−3.46, P = 2.31 × 10−2) are the significantly independent predictors of a shorter OS (Fig. 5D and E; Supplementary Fig. S9F; Supplementary Table S17).
Figure 5.
Prognostic impacts of genetic lesions. A and B, OS distributions in AEL and non-AEL (A) and PEL and EML (B) divided by TP53 mutation status. C, Kaplan–Meier survival curves are shown according to genetically clustered groups in the AEL cohort (A−D; Fig. 2). D, Kaplan–Meier survival curves are shown according to the presence or absence of EPOR gains/focal amplifications in TP53-mutated AEL. E, The result of multivariate analysis.
The negative effect of gains and/or focal amplifications on the EPOR locus within TP53-mutated AEL cases was also observed in the external cohort (18), although the platform used in the validation cohort failed to detect focal amplifications of EPOR and a substantial reduction of the statistical power was expected (Supplementary Fig. S9G and S9H).
Therapeutic Role of JAK2 Inhibition
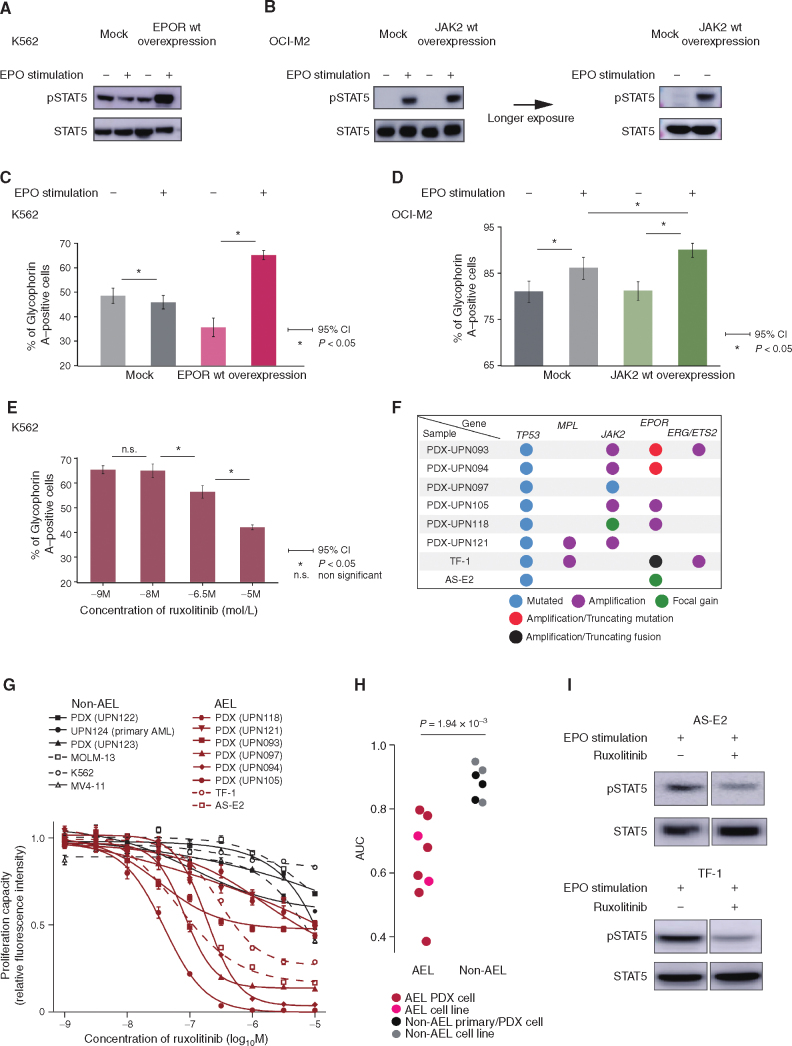
On the basis of frequent gains/focal amplifications involving EPOR/JAK2 loci and STAT5 activation in AEL, we overexpressed EPOR and JAK2 in K562 and OCI-M2 cell lines using lentivirus-mediated gene transfer, respectively, and evaluated the effect of the overexpression on STAT5 activation and erythroid differentiation in terms of glycophorin A (GPA) expression. We chose these cell lines because K562 is known to have high expression of JAK2 (37), and OCI-M2 has a focal amplification and overexpression of EPOR (ref. 37; Supplementary Fig. S10A). When stimulated with erythropoietin, EPOR-transduced K562 cells and OCI-M2 cells with and without JAK2 overexpression showed enhanced STAT5 phosphorylation (Fig. 6A and B) and upregulated GPA expression (Fig. 6C and D). Moreover, erythropoietin-induced GPA expression in K562 cells was suppressed by ruxolitinib in a dose-dependent manner (Fig. 6E). Taken together, we conclude that EPOR/JAK2 amplification contributes to STAT5 upregulation and erythroid phenotype of AEL cells having EPOR/JAK2 amplifications.
Figure 6.
EPOR–JAK2 signaling induces STAT5 activation and erythroid differentiation, and ruxolitinib suppresses the extent of proliferation and tyrosine phosphorylation of STAT5 induced by erythropoietin. A and B, Representative Western blot analysis for pSTAT5 expression in K562 cells with mock or EPOR overexpression (A) and OCI-M2 cells with mock or JAK2 overexpression (B) with or without erythropoietin (EPO) stimulation. B, The result of longer exposure of lysates without EPO stimulation is shown (right). C and D, The positivity of glycophorin A in K562 cells with mock or EPOR overexpression (C) or OCI-M2 with mock or JAK2 overexpression (D) cultured with and without EPO. E, The positivity of glycophorin A of K562 with EPOR overexpression cultured in RPMI-1640 with EPO under various ruxolitinib concentrations. F, Mutational status of TP53, MPL, JAK2, and ERG/ETS2 in AEL PDX and cell lines. Each colored dot represents a different mutational status. Focal gain is defined as CN gain spanning <107 base pair regions, and amplification is defined as a >2-fold increase (TCN > 4). G and H,In vitro effects of ruxolitinib on the proliferation capacity of AEL (red) and non-AEL (black) primary/PDX deprived cells (solid) and cell line (dash; G). Dot plots show area under the curve (AUC) of ruxolitinib which are compared between AEL cells (from 6 PDX models and 2 AEL cell lines; TF-1 and AS-E2) and non-AEL cells (from a viable cell stock, 2 PDX models, and 3 non-AEL cell lines; MOLM-13, K562, and MV4-11). Data analyzed by the Wilcoxon rank-sum test (H). I, Representative Western blot results (experiments were performed in triplicate). In vivo effects of ruxolitinib on the JAK/STAT pathway in AEL cell lines.
The functional relevance of frequent gains and/or focal amplifications involving JAK and/or EPOR and consequent STAT5 activation as shown above prompted us to test a possible therapeutic role of JAK2 inhibition for AEL cases carrying these genetic lesions. For this purpose, we newly established six PDXs from TP53-mutated AEL patients harboring gains/focal amplifications of JAK2/EPOR (PDX-UPN093, PDX-UPN094, PDX-UPN097, PDX-UPN105, PDX-UPN118, and PDX-UPN121) and tested their sensitivity to ruxolitinib in vitro, also including two publicly available AEL-derived cell lines (AS-E2, ref. 38; TF-1, ref. 39) carrying mutated TP53 and gains/amplifications affecting the EPOR and/or JAK2 loci together with a non-AEL primary sample, PDXs, and cell lines (Fig. 6F; Supplementary Fig. S10B–S10I). No gains or amplifications of EPOR or JAK2 were detected in non-AEL controls. When treated with different doses of ruxolitinib in vitro, all AEL-derived PDX cells and cell lines showed a higher response to JAK2 inhibition compared with non-AEL cells, based on significantly smaller area under the curve (AUC; Fig. 6G and H). In accordance with this, downregulated STAT5 phosphorylation (pSTAT5) was observed in ruxolitinib-treated AS-E2 and TF-1 cells in association with growth inhibition (Fig. 6I), supporting their dependence on activated JAK/STAT signaling.
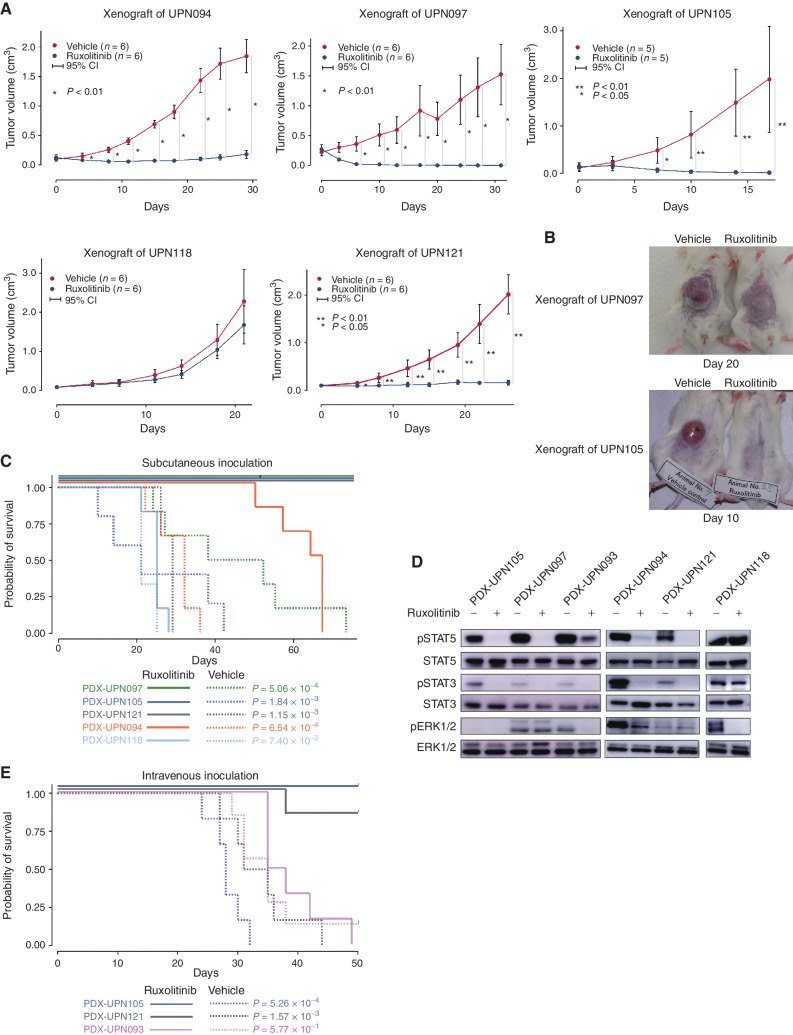
We also investigated the therapeutic effects of JAK2 inhibition using in vivo xenograft models treated with ruxolitinib, in which six AEL-derived PDXs were transplanted into immunodeficient NOD/SCID/γC-null (NOG; ref. 40) mice subcutaneously or intravenously, followed by 90 mg/kg ruxolitinib or a vehicle twice daily for 50 days after engraftment. Subcutaneous tumor growth was dramatically suppressed by ruxolitinib treatment in four (PDX-UPN094, PDX-UPN121 PDX-UPN105, and PDX-UPN121) of the five PDXs inoculated subcutaneously, where a prominent suppression of tumor growth (Fig. 7A and B) and a significantly prolonged survival (Fig. 7C) were observed. In agreement with this, an almost complete suppression of STAT5 phosphorylation was observed in tumor cells from ruxolitinib-treated mice transplanted in these four PDXs 6 hours after ruxolitinib treatment (Fig. 7D). Out of these four PDXs, two (PDX-UPN105 and PDX-UPN121) were also transplantable via intravenous inoculation, in which a substantial prolongation of survival was obtained with ruxolitinib treatment (Fig. 7E). By contrast, no growth suppression or prolongation of survival was observed in another subcutaneous model (PDX-UPN118; Fig. 7C). No prolongation of survival was obtained in PDX-UPN093 (transplantable only with intravenous inoculation) either (Fig. 7E). In these two ruxolitinib-resistant lines, there was no (PDX-UPN118) or only partial (PDX-UPN093) reduction of pSTAT5, suggesting a close link between tumor suppression and the suppression of STAT5 signaling on ruxolitinib treatment (Fig. 7D). PDX-UPN093 carried a highly amplified EPOR locus with a well-known activating mutation (p.G418X; Fig. 3D; Supplementary Fig. S10D). Moreover, another activating mutation (p.A364fs), although not amplified, was also seen in PDX-UPN094 (Fig. 3D; Supplementary Fig. S10E), which showed a slightly elevated pSTAT5 (Fig. 7D). Of note, all mice transplanted with PDX-UPN094 eventually died due to breakthrough tumor growth, despite a good initial response and a significantly prolonged survival. This suggests a possible role of concomitant activating EPOR mutations in resistance to ruxolitinib, although the mechanism of ruxolitinib resistance in the remaining PDX (PDX-UPN118) was still unknown with the lack of any accompanying mutations in the JAK/STAT signaling pathway. Taken together, these results suggest that inhibition of the JAK/STAT pathway might be a promising therapeutic strategy at least for a subset of TP53-mutated AEL with EPOR/JAK2 gains/focal amplification, although some cases do show ruxolitinib resistance.
Figure 7.
In vivo effects of ruxolitinib on AEL PDX cells. A–C,In vivo effects of ruxolitinib on AEL PDX cells. Oral administration of ruxolitinib (90 mg/kg twice a day) or vehicle control started after tumor formation of subcutaneously transplanted PDX cells into NOG immunodeficient mice for 50 days. Size of subcutaneous tumors (A), representative photos (B), and overall survivals (C) of indicated models were compared between ruxolitinib and vehicle control groups. C, Kaplan–Meier plots of subcutaneously inoculated models were shown. The effect of ruxolitinib on tumor size of each time point was analyzed using the Wilcoxon rank-sum test. The prognostic impact of ruxolitinib on OS was calculated by log-rank test. D, Representative Western blot results (experiments were performed in triplicate). In vivo effects of ruxolitinib on the JAK/STAT pathway in AEL PDX cells. Paired PDX cells of AEL PDX models treated once with ruxolitinib (90 mg/kg) vs. vehicle control (0.5% methyl cellulose) were obtained 6 hours after the treatment. Immunoblot shows the phosphorylation status of STAT3/5 and ERK1/2 in the AEL PDX models. E, Kaplan–Meier plots of intravenously inoculated models were shown. The prognostic impact of ruxolitinib on OS was calculated by the log-rank test.
DISCUSSION
Efforts to elucidate the unique pathophysiology of AEL based on extensive genome sequencing have successfully cataloged common genetic lesions in AEL (13–16), which has led to the identification of discrete genetic subclasses of AEL by Iacobucci and colleagues that are characterized by biallelic TP53 mutations, STAG2 and/or KMT2A-involving alterations, NPM1 mutations, DDX41 mutations, NUP98 rearrangements, and other lesions (18). We also confirmed these AEL subclasses together with their comutation patterns and impacts on survivals in our adult AEL cohort. Exceptions were DDX41-mutated and NUP98-fusion+ subclasses, to which only one each case belonged in our cohort (Fig. 2). This was anticipated because of the rarity of DDX41-mutated AEL cases and that the NUP98 fusion is highly specific to pediatric cases. However, we did confirm significantly elevated erythroblast counts in DDX41-mutated non-AEL cases compared with unmutated cases. We summarize similarities and differences in the results between the two studies in Supplementary Table S18.
However, despite the identification of major driver alterations and AEL subclasses, the underlying genetic lesions that can explain the unique phenotype of abnormal erythroid proliferation in AEL have long remained to be elucidated in previous studies. Thus, the identification of frequent gains/amplifications affecting genes for EPOR/MPL and their common downstream JAK2 signaling represents one of the major advances in the current study, given their undoubted functional link to abnormal erythroid proliferation (31, 41). Highly specific to TP53-mutated AEL cases with complex karyotypes with or without chromothripsis, EPOR/JAK2-involving SVs and CN lesions, together with those affecting ERG/ETS2, are thought to be causatively related to genetic instability associated with biallelic TP53 mutation. In particular, EPOR/JAK2-affecting gains/amplifications were highly enriched in PEL cases, compared with EML cases (10 or 77% of 13 PEL vs. 13 or 16% of 82 EML; odds ratio = 16.9; 95% CI, 3.9–80.7), which are now defined as genuine AEL by the presence of >80% of erythroblasts and >30% proerythroblast in BM in the current WHO classification (WHO 2017; ref. 11). The strong phenotype–genotype link between EPOR gains/amplifications and marked erythroid hyperplasia may lead to the definition of a novel category of “EPOR-amplified myeloid neoplasm.”
Also implicated in erythroid proliferation and/or differentiation (42–47), gains/amplifications of ERG/ETS2 were less specific to AEL and also found in non-AEL cases at a comparable frequency (Fig. 3G). However, when found in non-AEL, ERG/ETS2 lesions tend to be associated with increased erythroblast counts (P = 2.46 × 10−2). In addition, ERG/ETS2-affecting lesions were more common in TP53-mutated PEL (7/12 or 58%) than TP53-mutated EML (11/29 or 38%) cases, in which highly associated with EPOR gains/amplifications (Fig. 3E and G). Thus, ERG/ETS2 lesions still seem to contribute to the hypererythroid phenotypes in AEL. By contrast, the mechanism of erythroid proliferation in the remaining AEL cases is largely unclear. Given that an enhanced expression of STAT5 target genes was a common finding in AEL regardless of subgroup or genotype, abnormal erythroid proliferation in AEL could still be explained by activation of the JAK/STAT5 signaling pathway. In this regard, a strong correlation between STAG2 mutations and KMT2A-PTD, which characterizes group C AEL cases, might provide insight into the mechanism of aberrant erythroid proliferation. Another lesion of potential interest is USP9X mutation recurrently found in a subset of group D cases, because USP9X has previously been reported to suppress the JAK/STAT pathway (48, 49). However, the exact mechanism of aberrant erythroid proliferation in other AEL cases is largely unclear, and further functional studies should be warranted.
Another major finding in our study is a possible role of JAK2 inhibition in the therapeutics of AEL cases with EPOR gains/amplifications, which have an especially poor prognosis, compared even with other TP53-mutated AEL and non-AEL cases. Given the extremely dismal clinical outcomes of EPOR-amplified AEL cases, the efficacy of JAK2 kinase inhibition is worthwhile testing for these cases in a clinical setting. Moreover, the uniform activation of STAT5 in AELs may predict a role of JAK2 inhibition in other subtypes of AEL, including group B to D cases. Nevertheless, we did observe ruxolitinib resistance in some PDX cases, where an activating EPOR mutation was implicated. Further investigations are required to confirm the effect of ruxolitinib in clinical settings and to elucidate the exact mechanism of ruxolitinib resistance.
Finally, despite a clear correlation between gains/amplifications/mutations of EPOR/JAK2, the mechanism of the characteristic erythroid-dominant phenotype is still unclear for other AEL subclasses, where the majority of cases are EML, and the erythroid proliferation is less conspicuous than PEL. While sharing common class-defining mutations (i.e., mutated TP53 with aneuploidy, NPM1, or STAG2 mutation), each genetic AEL subtype differs from the corresponding non-AEL counterpart with regard to comutation patterns, which therefore might explain the erythroid-dominant phenotype of AEL. For example, gains/amplifications of EPOR/JAK2 in TP53-mutated cases, KMT2A-PTD in STAG2-mutated cases, and underrepresentation of FLT3-ITD and mutations and overrepresentation of PTPN11 in NPM1-mutated AEL might be of interest, together with recurrent USP9X mutations in TN cases. Elucidation of the mechanistic basis of their AEL phenotype in these subclasses is among the major challenges in further investigation.
METHODS
Patients
We collected 121 patients with AEL as per the criteria proposed by the WHO (WHO 2001; ref. 26), i.e., more than 80% erythroblasts (pure erythroid leukemia; PEL) or more than 50% of erythroblasts together with >20% blasts (erythroid/myeloid leukemia; EML; Supplementary Table S19). They included 13 PEL, 82 EML, and other 26 cases, whose diagnostic details were unknown. Note that according to the most updated WHO classification (WHO 2017; ref. 11), the diagnosis in 61 EML cases should be revised to MDS-EB. We also included 214 cases with non-AEL based on the same WHO 2001 criteria who had been enrolled at our collaborating institutes between July 1, 2017, and July 31, 2019, and agreed to participate in this study (Supplementary Tables S20 and S18). All participants provided written informed consent. In addition, we used the data set of major driver mutations and CNAs obtained from the targeted-capture sequencing in 229 cases with MDS-EB (in WHO 2017) without erythroid hyperplasia to elucidate the difference in genetic profiles between AEL with <20% total blasts and MDS-EB without erythroid hyperplasia (see also below). These MDS-EB cases without erythroid hyperplasia were a subset of a larger cohort of MDS cases used in the previous study (50) and selected from the two different cohorts, including those from the JALSG MDS212 trial (51) and our own biobank at Kyoto University, which had been consecutively collected for studies on different topics from January 2013 to June 2018 and July 2017 to June 2021, respectively. All samples analyzed in the study were obtained according to the protocols by the ethics board of each participating institution.
TCGA-LAML Data Set
In addition to these “in-house” cases, we also included the data set of the TCGA-LAML project (dbGaP Study Accession: phs000178.v11.p8; ref. 7): published cases of 3 AEL and 195 non-AEL cases as per the WHO 2001 criteria, for which WES as well as RNA (2 AEL and 175 non-AEL) sequencing data were available from TCGA (ref. 7; Supplementary Table S1). Two cases with a diagnosis of FAB classification were unavailable and were omitted from the analysis. Bam files were obtained and analyzed using Genomon 2.
This study was conducted in accordance with the Declaration of Helsinki and has been approved by the Ethics Committee of the Faculty of Medicine, Kyoto University.
In total, we included 124 AEL, 409 non-AEL, and 229 MDS-EB cases without erythroid hyperplasia in the current study.
The median age at enrollment of AEL, non-AEL, and MDS-EB cohorts was similar: 60 (21–87.1), 59 (17–90), and 72 (16–92) years, respectively. As for sex, the frequency of males is higher in AEL (74.2%) and MDS-EB (73%) without erythroid hyperplasia than in non-AEL AML (56.1%).
Power Analysis
No statistical methods were used to predetermine sample size. As the main aim of this study was to explore the genomic profile of AEL, we collected as many samples as possible.
Randomization
This study was performed in order to clarify the difference between AEL and non-AEL and between AEL and MDS-EB. Patients were divided according to diagnostic criteria. Thus, randomization was not performed.
As the aim of this study was to clarify the difference between AEL and non-AEL and between AEL and MDS-EB, blinding was not performed.
WGS/WES
We analyzed paired tumor and germline DNA from 35 AEL patients, including 6 PEL, 16 EML, and 13 other AEL cases, using WGS (n = 20) and/or WES (n = 27), which were performed as previously described (29, 52). Briefly, tumor and germline DNA was extracted from patients’ BM or peripheral blood mononuclear cells and from buccal mucosa, respectively, using the QIAamp DNA Mini Kit (QIAGEN, cat. #51304) according to the manufacturer's instructions. Samples were subjected to massively parallel sequencing with 150 bp paired-end reads using the HiSeq 2000, HiSeq2500, HiSeq X Ten, and/or NovaSeq 6000 according to the manufacturer's instructions. Sequencing reads were aligned to NCBI Human Reference Genome Build 37 (hg19) by Burrows−Wheeler Aligner, version 0.7.10, with default parameters (http://bio-bwa.sourceforge.net/). PCR duplicates were eliminated using Picard tools version 1.39 (GATK). Mutation calling was performed using the Empirical Bayesian Mutation Calling (EBCall) algorithm (53) with the following parameters:
(i) Mapping quality score ≥20
(ii) Base quality score ≥15
(iii) Both tumor and normal depths ≥8
(iv) Number of variant reads in tumors ≥4
(v) VAFs in tumor samples ≥0.05
(vi) VAFs in normal samples ≤0.2.
We used stringent criteria for mutation calling, requiring a P value (by EBCall) <10−4 and a Fisher P < 10−1.3, as determined by counting the number of reads with the reference base and the candidate single-nucleotide variant (SNV) and short insertion/deletion (in/del) in both the tumor and normal samples as validated mutations.
The number of SV events of each sample analyzed by WGS is calculated using ClusterSV (54). Candidate mutations were filtered in the same manner as for WES analysis and included the following additional criteria.
(i) Mutations with ≥3 variant reads in tumor samples
(ii) Synonymous SNVs and
(iii) The exclusion of mutations with P ≥ 0.001 (EBCall).
Detection of structural variations was performed by Genomon SV as previously reported (55, 56). Briefly, Genomon SV used the information from chimeric reads (containing breakpoints) and discordant read pairs, and reads were aligned to the assembled contig sequence containing the SV breakpoint (variant sequence) for each candidate SV. The Fisher exact test compared the proportion of the read pairs aligned to variant sequences relative to the reference sequences in tumor versus matched normal samples. Putative SVs were manually curated and filtered by removing those with
(i) Fisher exact P > 0.1;
(ii) <4 supporting reads in tumor samples;
(iii) <0.05 variant allele frequency (VAF) in tumor samples;
(iv) ≥0.02 VAF in matched normal samples; or
(v) <1,000 bp distance between breakpoints.
Targeted-Capture Sequencing
Subsequently, a total of 121 cases with AEL were screened for mutations in 376 genes (Supplementary Table S3) associated with myeloid neoplasms (7, 8, 19, 27–29), erythroid differentiation process, and 1,216 SNP sites for CN detection (20) by targeted-capture sequencing as previously described (27, 57). Briefly, DNA was enriched for target exons by liquid phase hybridization using the SureSelect custom kit (Agilent Technology). Sequencing reads were aligned as described for WGS/WES. Mutation calling was performed as previously reported (27, 57). Briefly, mutation calling was performed using our established pipeline Genomon 2 (http://genomon-project.github.io/GenomonPages/), as previously reported (27, 57) using the following inclusion and exclusion parameters:
The candidates with the following criteria were included:
(i) Mapping quality score ≥20
(ii) Base quality score ≥15
(iii) Number of SNVs on the same read <5
(iv) Number of insertions and deletions on the same read <2
(v) Number of total reads ≥20
(vi) Number of variant reads ≥4
(vii) VAFs ≥0.02
The candidates with the following criteria were excluded:
(i) Synonymous and ambiguous (unknown) variants
(ii) Variants that read only from one direction
(iii) Single-nucleotide substitutions in which other mutations were called at the same position and their VAFs were ≥0.1.
Further, SVs were called using the in-house pipeline Genomon SV (55, 56). Finally, candidates that fulfilled all the following criteria were adopted:
(i) The contig sequence aligned to the nucleotides to the left and right of the SV breakpoint pairs (maximum overhang ≥65 bp)
(ii) The contig sequence aligned to the coding region of targeted genes
(iii) They were not called in normal control samples
(iv) They had an allele frequency ≥0.05.
Finally, mapping errors were removed by visual inspection on the Integrative Genomics Viewer (IGV) browser (http://software.broadinstitute.org/software/igv/). For 3 samples from 3 patients, amplified DNA was used for sequencing analysis.
Curation of the Oncogenic Variants
The detected candidate variations fulfilling the quality filter noted above were assumed to be “oncogenic” and were included in the subsequent analyses when these variants fulfilled one of the following criteria:
(i) Candidates that were registered in the Catalog of Somatic Mutations in Cancer (COSMIC) v70 database ≥5 times in whole cancer tissues and/or ≥1 in the hematopoietic and lymphoid tissues at the given genomic positions and base substitutions.
(ii) Candidates that fulfill all Criteria 1 and at least one of the Criteria 2.
Criteria 1
a. Candidates whose VAFs were ≥0.03.
b. Candidates who were not registered in our in-house polymorphism database, NCBI dbSNP Build 131, or other public databases (minor allele frequency >0.001), including ESP6500, the 1000 Genomes Project as of April 23, 2012, the Human Genome Variation Database, and the ExAC database.
c. EBCall >4.
Criteria 2
a. Candidates who were located on the nonrepeat region with VAFs ≥0.03 <0.4 or ≥0.6 <0.98.
b. Nonsense, frameshift, and splice-site candidates with VAFs ≥0.03.
Estimation of Tumor Cell Fractions
The tumor cell fraction (TCF) was estimated from the total copy-number (TCN) of the region and the minor allele-specific CN (AsCN) using the following formula:
The estimated TCF harboring the relevant mutation was calculated using the TCN of the region and the observed VAF value, as previously described.
Copy number of gains of tumor cell was calculated as follows (57):
Copy-Number Analysis
In addition to the evaluation of the conventional metaphase karyotyping, we developed a novel sequencing−based platform for copy−number analysis, named CNACS (58), which quantifies total copy numbers and allele-specific copy numbers based on sequencing depths and allelic ratios. Correction for multiple biases in CN signals allowed for higher resolution. By applying CNACS to sequencing data of patients’ genomic DNA, we detected copy-number changes and copy-neutral LOH mostly caused by uniparental disomy (UPD). In cases examined by CNACS, focal gain is defined as copy-number gain spanning less than 107 base pair regions, and amplification is defined as greater than a 2-fold increase (TCN >4). For CN analysis of WGS data, we applied the Control-FREEC algorithm as previously described (59).
RNA Sequencing
RNA sequencing was performed as previously described (58). Briefly, total RNA was extracted from the whole bone marrow (n = 59) of 21 AEL patients and 38 non-AEL patients using the RNeasy Micro Kit (QIAGEN; cat. #74004) according to the manufacturer's instructions (Supplementary Table S1). RNA sequencing libraries were prepared from polyA-selected RNA using the NEBNext Ultra RNA Library Prep kit for Illumina (New England BioLabs; cat. #E7370). Libraries were sequenced using the Illumina HiSeq 2500 platform with a standard 100 bp paired-end read protocol. Alignment to the human reference genome (hg19) and fusion detection was conducted by Genomon v2.6.3, using the following criteria:
- (i) At least three spanning reads.
- (ii) Junctions located at known exon–intron boundaries.
- (iii) Fusion transcripts of two different genes.
All genomic coordinates are based on GRCh37/hg19. For expression analysis, mapped reads were counted for each gene by our in-house Genomon Expression pipeline (http://github.com/Genomon-Project/GenomonExpression). Gene-expression normalization and differential expression analysis were performed using the Bioconductor package DESeq2 (60). or gene set enrichment scores, weighted Kolmogorov–Smirnov-like statistics were estimated, and empirical permutation tests by shuffling group labels of the samples were performed to evaluate the significance of enrichment scores. Gene sets with q < 0.1 were considered significantly enriched.
Cell Lines
The cell line AS-E2 was generously provided by the originator, Yasushi Miyazaki (Nagasaki University). The other cell lines (K562, MOLM13, MV4-11, TF-1, OCI-M2, and 293T) were obtained from the ATCC. None of the cell lines used were authenticated. K562 and MOLM13 were verified as Mycoplasma spp. negative using MycoALERT (Lonza; cat. #LT07-218). The other cell lines were not tested. Experiments using cell lines were performed 1 week after thawing. We overexpressed EPOR and JAK2 in K562 and OCI-M2 cell lines using lentivirus-mediated gene transduction. For the generation of lentiviruses, 293T cells were transfected with the gene overexpression constructs, psPAX2 (Addgene; cat. #12260) and pMD2.G plasmid (Addgene; cat. #12259). Lentivirus for JAK2/EPOR overexpression was constructed by inserting JAK2/EPOR cDNA into the MCS of the CSII-EF backbone vector, which was provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED. Transfections in 293T cells were performed using Polyethylenimine MAX (Polysciences; cat. #24765-1) reagent at 4:3:1 ratios of vector: psPAX2: pMD2.G in OPTI-MEM solution (Thermo Fisher Scientific; cat. #31985070). Viral supernatant was collected 36 hours and 48 hours after transfection and subjected to ultracentrifugation (20,000 × g for 5 hours.) to concentrate lentiviral particles. Each cDNA was synthesized at Eurofins Genomics K.K. The sequences of insert cDNA were provided in Supplementary Table S21. Spin infections were performed at room temperature at 1,200 × g for 120 minutes with polybrene reagent (Thermo Fisher Scientific; cat. #TR1003G) at a final concentration of 4 μg/mL.
Establishment of Xenograft Mouse Models
Animal care was in accordance with institutional guidelines and approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University (Kyoto, Japan). Patient-derived xenograft (PDX) models were established by injecting bone marrow or peripheral blood mononucleated cells of AML patients into newborn NOG (NOD/SCID/IL2rγnull) mice (40), which were purchased from the Central Institute for Experimental Animals (Kawasaki, Japan). We also obtained PDX models from PRoXe (61).
In Vivo Drug Efficacy Test Using PDX Mouse Models
Six-week-old female NOD/SCID/γC-null (NOG) mice were used for drug efficacy tests. Animal care was in accordance with institutional guidelines and approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University (Kyoto, Japan). After confirming tumor engraftment, ruxolitinib (LC Laboratories; cat. #R-6600) or vehicle was given twice daily dose (90 mg/kg) via oral gavage for 50 days.
Inhibitor Assay
PDX cells and cell lines were plated at a density of 2–10 × 105 cells/mL in 96-well plates (50 μL per well) with ruxolitinib or a vehicle (dimethyl sulfoxide, DMSO; Agilent, cat. #600260-53). After culturing in Iscove's modified Dulbecco's medium (IMDM; FUJIFILM Wako Pure Chemical Corporation; cat. #098-06465) with 2U human EPO (PeproTech; cat. #100-64), 20% FBS (Nichirei Biosciences; cat. #175012), and 1% penicillin–streptomycin (FUJIFILM Wako Pure Chemical Corporation; cat. #168-23191) at 37°C and 5% CO2 for 3 days. Cell viability was assessed by quantification of ATP using CellTiter-Glo (Promega; cat. #G9242). The 4-parameter logistic regression models were used to assess the inhibitory effect of ruxolitinib using Prism software (v6; GraphPad).
Immunoblotting
For the preparation of lysates, cells were harvested and lysed with RIPA buffer (Nacalai Tesque; cat. #16488-34) containing 50 mmol/L Tris-HCL buffer, 150 mmol/L sodium chloride, 1% Nonidet(R) P40 substitute, 0.5% deoxycholic acid sodium salt monohydrate, and 0.1% sodium dodecyl sulfate, supplemented with 1 × phosphatase inhibitor cocktail (Nacalai Tesque; cat. #07574-61) at 4°C for 30 minutes with constant vortex every 3–5 minutes. The lysate was transferred to a microcentrifuge tube and centrifuged at ∼150,000 rpm for 10 minutes at 4°C, and the supernatant was transferred to a new microcentrifuge tube and discard the pellet. After boiled in Sample Buffer Solution with Reducing Reagent (6×) for SDS-PAGE (Nacalai Tesque; cat. #09499-14) at 95°C for 5 minutes, cell lysates were analyzed by western blotting using the following antibodies, which were purchased from Cell Signaling Technology and used at a 1:1,000 dilution: anti-tyrosine phosphorylation (p) of STAT5A (cat. 4322S), anti-STAT5 (9310), anti−p-p44/42 ERK1/2 (4370S), anti-p44/42 ERK1/2 (4695S), anti-pSTAT3 (9145), and anti-STAT3 (4904). Multiple independent blots were used, and for the blots of one experiment, the same amount of lysate was applied for each sample. For cell lines, TF-1 and AS-E2 cells were treated with 2 U/mL erythropoietin (EPO (PeproTech; cat. #100-64)) for 10 minutes after serum starvation (1%) for 16 hours and exposure of 10−3 mol/L ruxolitinib or vehicle (DMSO; Agilent; cat. #600260-53) for 3 hours.
Measurement of Glycophorin A Expression
K562 were cultured in RPMI-1640 (Nacalai tesque; cat. #30264-56) with 10% FBS (Nichirei Biosciences; cat. #175012) with or without 2 U/mL EPO (PeproTech; cat. #100-64), and OCI-M2 were cultured in IMDM (FUJIFILM Wako Pure Chemical Corporation; cat. #098-06465) with 1% FBS with or without 1 U/mL EPO for 3 days. Percentages of glycophorin A–positive cells were determined by glycophorin A–APC antibody [BD Biosciences; cat. #551336 clone: GA-R2 (HIR2)].
RT-qPCR
Reverse transcription of RNA was performed using the random hexamer and ReverTra Ace qPCR RT Kit (TOYOBO; cat. #FSQ-301). The PCR amplicon is designed to span over intron regions using NCBI Primer-BLAST. Quantitative PCR was performed in triplicate using TB Green Premix Ex Taq II (Takara; cat. #RR820A) on a CFX Connect Real-Time PCR Detection System. The detailed primer sequences for RT-qPCR are provided as follows:
GAPDH forward: 5′-AACGTGTCAGTGGTGGACCTG-3′; reverse: 5′-AGTGGGTGTCGCTGTTGAAGT-3′
JAK2 forward: 5′-CACCTAAGAGACTTTGAAAGGGAAA-3′; reverse: 5′-TTAGATTACGCCGACCAGCA-3′
EPOR forward: 5′-CGTATGGCTGAGCCGAGCTT-3′; reverse: 5′-AGCACCAGGATGACCACGA-3′
ERG forward: 5′-CAACAAGTAGCCGCCTTGC-3′; reverse: 5′-ATCTTGAACTCCCCGTTGGT-3′
ETS2 forward: 5′-GGAATCAAGAATATGGACCAGGTAG-3′; reverse: 5′-ACCCATCAAAGGTGTCAAAGG-3′
MPLforward: 5′-GGTGAAGAATGTGTTCCTAAACCAG-3′; reverse: 5′-CTCCTCCCAGCTGATCTGAAGT-3′
Statistical Analysis
Data are expressed as a mean ± 95% confidence interval unless otherwise indicated. Pairwise comparisons were performed using the Wilcoxon rank-sum test for continuous variables and the two-sided Fisher exact test for categorical variables. Where zeros cause problems with the computation of the odds ratio, 0.5 is added to all values. The Kaplan–Meier method was used to analyze survival outcomes (OS) using the log-rank test or Cox proportional hazards model.
Multivariable analysis of OS was performed including 48 cases in group A, in which survival data were available, based on the Cox proportional hazard model using the backward stepwise selection for variable selection. In addition to age and sex, we included all the mutations and CNAs in the multivariable analysis, which were observed in >20% of group A cases and significant in univariate analysis (P < 0.05), i.e., age, sex, and gains/amplifications of EPOR locus on 19p (Supplementary Fig. S9F). All statistical analyses were performed using the R (http://www.R-project.org) or STATA/IC (LightStone) ver. 13.1. Significance was determined at a two-sided α-level of 0.05, except for P values in multiple comparisons, in which multiple tests were adjusted according to the method described by Benjamini and Hochberg (62). Methods of detailed statistical analyses are described in each section above.
Data Availability
Data sets of sequencing in samples with AEL are available in the European Genome−phenome Archive database (Accession ID: EGAS00001003696 and EGAS00001005810).
Supplementary Material
Acknowledgments
We thank Atsuko Ryu, Maki Nakamura, Takeshi Shirahari, Kazuhide Onishi, Kumiko Sato, and Natsumi Sakamoto (Department of Pathology and Tumor Biology, Kyoto University) for technical assistance. This work was supported by grants from the Japan Agency for Medical Research and Development (no. 19ck0106470h0001 to H. Makishima; and nos. JP15cm0106056h0005, JP19cm0106501h0004, JP16ck0106073h0003, and JP19ck0106250h0003 to S. Ogawa); the Ministry of Education, Culture, Sports, Science and Technology of Japan; the High Performance Computing Infrastructure System Research Project (nos. hp160219, hp170227, hp180198, and hp190158 to S. Ogawa). (This research used computational resources of the K computer provided by the RIKEN Advanced Institute for Computational Science through the HPCI System Research project); the Japan Society for the Promotion of Science; Scientific Research on Innovative Areas (no. JP15H05909 to S. Ogawa); KAKENHI (nos. JP26221308 and JP19H05656 to S. Ogawa, JP19H01053 to H. Makishima, and 22H02817 to A. Yoda); and the Takeda Science Foundation to S. Ogawa. S. Ogawa is a recipient of the JSPS Core-to-Core Program A: Advanced Research Networks. We thank the Central Institute for Experimental Animals (Kawasaki, Japan) for providing NOG mice, and the TCGA Consortium and all its members for making publicly available their invaluable data.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
M.M. Nakagawa reports grants from Sumitomo-Dainippon Pharma Co., Ltd. outside the submitted work. Y. Nannya reports grants from Japan Agency for Medical Research and Development and KAKENHI during the conduct of the study; personal fees from Otsuka Pharmaceuticals, Takeda Pharmaceuticals, Pfizer, Chugai Pharmaceuticals, Dai Nippon Sumitomo, Astra Zeneca, Novartis, Kyowa-Hakko Kirin, Fuji-Seiyaku, Asahi-Kasei, Nippon Shinyaku, and Bristol-Myers outside the submitted work. A. Yoda reports grants, personal fees, and nonfinancial support from Chordia Therapeutics Inc. during the conduct of the study; grants, personal fees, and nonfinancial support from Chordia Therapeutics Inc. outside the submitted work. Y. Ochi reports grants from Nippon Shinyaku Co., Ltd outside the submitted work. D. Morishita reports other support from Chordia Therapeutics Inc. outside the submitted work. A. Hangaishi reports grants from Takeda Pharmaceutical Company, Kyowa Kirin, and personal fees from Chugai Pharmaceutical Co outside the submitted work. S. Miyano reports grants from the Ministry of Education, Science, Sports and Culture, Japan during the conduct of the study; nonfinancial support from Liquid Mine Ltd. and personal fees from Fujitsu Ltd. outside the submitted work. S. Chiba reports grants from a government agency, academic society, commercial sponsors, and personal fees from commercial sponsors outside the submitted work. Y. Miyazaki reports grants from Sumitono Pharma, Chugai Pharmaceutica, personal fees from AbbVie, Novartis, Astellas, SymBio, Sumitomo Pharma, Bristlo Myers Squibb, Nippon Shinyaku, Otsuka Pharma, and Kyowa Kirin outside the submitted work. K. Usuki reports grants from Astellas-Amgen-Biopharma, grants and personal fees from Otsuka, SymBio, Takeda, Nippon Shinyaku, Novartis, AbbVie, Bristo-Myers Squibb, Ono, Chugai, Daiichi-Sankyo, MSD, Astellas, Alexion, Kyowa Kirin, Pfizer, Celgene, Yakult, Eisai, PharmaEssentia, grants from Apellis, Janssen, Gilead, Incyte, Sumitomo-Dainippon, and Mundi, personal fees from Sanofi and Alnylam Japan outside the submitted work. S. Miyawaki reports personal fees from Ohtsuka Pharmaceutical, Astelas, and Nippon Shinyaku Co. outside the submitted work. K. Ohyashiki reports other support from Celgene, Novartis, and Bristol-Myers Squibb outside the submitted work. M. Heuser reports grants and personal fees from AbbVie, Jazz Pharmaceuticals, Novartis, Daiichi-Sankyo, Glycostem, Pfizer, PinotBio, Roche, personal fees from Eurocept, Janssen, Takeda, Agios, BMS, Kura Oncology, Tolremo, grants from Astellas, Bayer Pharma AG, BergenBio, and LoxoOncology outside the submitted work. F. Thol reports other support from BMS and AbbVie outside the submitted work. A. Takaori-Kondo reports grants and personal fees from Astellas Pharma Inc., personal fees from Bristol-Myers Squibb Co, Janssen Pharmaceutical K.K., grants from Chugai Pharmaceutical Co., Ltd., DKS Co. Ltd., Eisai, the Japanese Society of Hematology, OHARA Pharmaceutical Co.,Ltd, Ono Pharma Inc., Sanofi K.K., SHIONOGI and Co., Ltd., and Takeda Pharmaceutical Company Limited, grants and personal fees from Kyowa Kirin Co., Ltd., Nippon Shinyaku Co., Ltd., Otsuka Pharmaceutical Co., Ltd., other support from Megakaryon Co., personal fees from Novartis Pharma K.K. outside the submitted work; in addition, A. Takaori-Kondo has a patent for COGNANO, Inc. pending and a patent for NextGem Inc. pending. S. Ogawa reports grants from the Japan Agency for Medical Research and Development, the Japan Society for the Promotion of Science, the Ministry of Education, Culture, Sports, Science and Technology, and Takeda Science Foundation during the conduct of the study; grants from Japan Agency for Medical Research and Development, Chordia Therapeutics, Japan Science and Technology Agenecy, personal fees from Novartis Pharmaceuticals and Astellas Pharmaceuticals outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
J. Takeda: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft. K. Yoshida: Conceptualization, data curation, supervision, investigation, visualization, methodology, writing–original draft. M.M. Nakagawa: Visualization. Y. Nannya: Data curation, supervision. A. Yoda: Resources, investigation. R. Saiki: Resources, visualization. Y. Ochi: Resources, investigation. L. Zhao: Resources. R. Okuda: Resources. X. Qi: Resources. T. Mori: Resources. A. Kon: Resources. K. Chiba: Methodology. H. Tanaka: Methodology. Y. Shiraishi: Methodology. M. Kuo: Resources. C.M. Kerr: Resources. Y. Nagata: Resources. D. Morishita: Methodology. N. Hiramoto: Resources. A. Hangaishi: Resources. H. Nakazawa: Resources. K. Ishiyama: Resources. S. Miyano: Methodology. S. Chiba: Resources. Y. Miyazaki: Resources. T. Kitano: Resources. K. Usuki: Resources. N. Sezaki: Resources. H. Tsurumi: Resources. S. Miyawaki: Resources. J.P. Maciejewski: Resources. T. Ishikawa: Resources. K. Ohyashiki: Resources. A. Ganser: Conceptualization, resources. M. Heuser: Conceptualization, resources. F. Thol: Conceptualization, resources. L. Shih: Resources. A. Takaori-Kondo: Resources. H. Makishima: Writing–original draft. S. Ogawa: Conceptualization, supervision, writing–original draft, project administration.
References
- 1. Schwartz SO, Critchlow J. Erythremic myelosis (Dl Guglielmo's disease): critical review with report of four cases, and comments on erythroleukemia. Blood 1952;7:765–93. [PubMed] [Google Scholar]
- 2. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol 1976;33:451–8. [DOI] [PubMed] [Google Scholar]
- 3. Garand R, Duchayne E, Blanchard D, Robillard N, Kuhlein E, Fenneteau O, et al. Minimally differentiated erythroleukaemia (AML M6 ‘variant’): a rare subset of AML distinct from AML M6. Groupe Francais d'Hematologie Cellulaire. Br J Haematol 1995;90:868–75. [DOI] [PubMed] [Google Scholar]
- 4. Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol 1999;17:3835–49. [DOI] [PubMed] [Google Scholar]
- 5. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009;114:937–51. [DOI] [PubMed] [Google Scholar]
- 6. Lessard M, Struski S, Leymarie V, Flandrin G, Lafage-Pochitaloff M, Mozziconacci MJ, et al. Cytogenetic study of 75 erythroleukemias. Cancer Genet Cytogenet 2005;163:113–22. [DOI] [PubMed] [Google Scholar]
- 7. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Copelli M. Di una emopatia sistemizzata rapresentata da una imperplasia eritroblastica (eritronatosis). Pathologica 1912;4:460. [Google Scholar]
- 10. Di Guglielmo G. Richerche di ematologia. I. Un caso di eritroleucemia. Megacariociti in circolo e loro funzione piastrinopoietico. Folia Medica (Pavia) 1917;13:386. [Google Scholar]
- 11. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 12. Boddu P, Benton CB, Wang W, Borthakur G, Khoury JD, Pemmaraju N. Erythroleukemia-historical perspectives and recent advances in diagnosis and management. Blood Rev 2018;32:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grossmann V, Bacher U, Haferlach C, Schnittger S, Potzinger F, Weissmann S, et al. Acute erythroid leukemia (AEL) can be separated into distinct prognostic subsets based on cytogenetic and molecular genetic characteristics. Leukemia 2013;27:1940–3. [DOI] [PubMed] [Google Scholar]
- 14. Cervera N, Carbuccia N, Garnier S, Guille A, Adelaide J, Murati A, et al. Molecular characterization of acute erythroid leukemia (M6-AML) using targeted next-generation sequencing. Leukemia 2016;30:966–70. [DOI] [PubMed] [Google Scholar]
- 15. Ping N, Sun A, Song Y, Wang Q, Yin J, Cheng W, et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia 2017;31:195–202. [DOI] [PubMed] [Google Scholar]
- 16. Montalban-Bravo G, Benton CB, Wang SA, Ravandi F, Kadia T, Cortes J, et al. More than 1 TP53 abnormality is a dominant characteristic of pure erythroid leukemia. Blood 2017;129:2584–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cervera N, Carbuccia N, Mozziconacci MJ, Adelaide J, Garnier S, Guille A, et al. Revisiting gene mutations and prognosis of ex-M6a-acute erythroid leukemia with regard to the new WHO classification. Blood Cancer J 2017;7:e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iacobucci I, Wen J, Meggendorfer M, Choi JK, Shi L, Pounds SB, et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet 2019;51:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 2020;26:1549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu W, Hasserjian RP, Hu Y, Zhang L, Miranda RN, Medeiros LJ, et al. Pure erythroid leukemia: a reassessment of the entity using the 2008 World Health Organization classification. Mod Pathol 2011;24:375–83. [DOI] [PubMed] [Google Scholar]
- 22. Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov 2019;9:910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ali D, Jönsson-Videsäter K, Deneberg S, Bengtzén S, Nahi H, Paul C, et al. APR-246 exhibits anti-leukemic activity and synergism with conventional chemotherapeutic drugs in acute myeloid leukemia cells. Eur J Haematol 2011;86:206–15. [DOI] [PubMed] [Google Scholar]
- 24. Iacobucci I, Qu C, Varotto E, Janke LJ, Yang X, Seth A, et al. Modeling and targeting of erythroleukemia by hematopoietic genome editing. Blood 2021;137:1628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shoshani O, Brunner SF, Yaeger R, Ly P, Nechemia-Arbely Y, Kim DH, et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 2021;591:137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002;100:2292–302. [DOI] [PubMed] [Google Scholar]
- 27. Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014;28:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013;122:3616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017;49:204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iacobucci I, Li Y, Roberts KG, Dobson SM, Kim JC, Payne-Turner D, et al. Truncating erythropoietin receptor rearrangements in acute lymphoblastic leukemia. Cancer Cell 2016;29:186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He TC, Jiang N, Zhuang H, Quelle DE, Wojchowski DM. The extended box 2 subdomain of erythropoietin receptor is nonessential for Jak2 activation yet critical for efficient mitogenesis in FDC-ER cells. J Biol Chem 1994;269:18291–4. [PubMed] [Google Scholar]
- 32. Ogawa S. Genetics of MDS. Blood 2019;133:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015;125:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 35. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. An X, Schulz VP, Li J, Wu K, Liu J, Xue F, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014;123:3466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miyazaki Y, Kuriyama K, Higuchi M, Tsushima H, Sohda H, Imai N, et al. Establishment and characterization of a new erythropoietin-dependent acute myeloid leukemia cell line, AS-E2. Leukemia 1997;11:1941–9. [DOI] [PubMed] [Google Scholar]
- 39. Kitamura T, Tojo A, Kuwaki T, Chiba S, Miyazono K, Urabe A, et al. Identification and analysis of human erythropoietin receptors on a factor-dependent cell line, TF-1. Blood 1989;73:375–80. [PubMed] [Google Scholar]
- 40. Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 2002;100:3175–82. [DOI] [PubMed] [Google Scholar]
- 41. Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, et al. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell 1993;74:227–36. [DOI] [PubMed] [Google Scholar]
- 42. Birger Y, Goldberg L, Chlon TM, Goldenson B, Muler I, Schiby G, et al. Perturbation of fetal hematopoiesis in a mouse model of Down syndrome's transient myeloproliferative disorder. Blood 2013;122:988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carmichael CL, Metcalf D, Henley KJ, Kruse EA, Di Rago L, Mifsud S, et al. Hematopoietic overexpression of the transcription factor Erg induces lymphoid and erythro-megakaryocytic leukemia. Proc Natl Acad Sci U S A 2012;109:15437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang JZ, Carmichael CL, Shi W, Metcalf D, Ng AP, Hyland CD, et al. Transposon mutagenesis reveals cooperation of ETS family transcription factors with signaling pathways in erythro-megakaryocytic leukemia. Proc Natl Acad Sci U S A 2013;110:6091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fagnan A, Bagger FO, Piqué-Borràs MR, Ignacimouttou C, Caulier A, Lopez CK, et al. Human erythroleukemia genetics and transcriptomes identify master transcription factors as functional disease drivers. Blood 2020;136:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ge Y, LaFiura KM, Dombkowski AA, Chen Q, Payton SG, Buck SA, et al. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia 2008;22:521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baldus CD, Liyanarachchi S, Mrózek K, Auer H, Tanner SM, Guimond M, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A 2004;101:3915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwartzman O, Savino AM, Gombert M, Palmi C, Cario G, Schrappe M, et al. Suppressors and activators of JAK-STAT signaling at diagnosis and relapse of acute lymphoblastic leukemia in Down syndrome. Proc Natl Acad Sci U S A 2017;114:E4030–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chou DH, Vetere A, Choudhary A, Scully SS, Schenone M, Tang A, et al. Kinase-independent small-molecule inhibition of JAK-STAT signaling. J Am Chem Soc 2015;137:7929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernard E, Tuechler H, Greenberg Peter L, Hasserjian Robert P, Arango Ossa Juan E, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evidence 2022;1:EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 51. Kiguchi T, Sato S, Usuki K, Ishiyama K, Ito Y, Suzuki T, et al. Prospective comparison of azacitidine treatment between 7-days and 5-days schedules for patients with higher-risk myelodysplastic syndromes; results of Japan Adult Leukemia Study Group MDS212 Trial. Blood 2019;134:845.31515225 [Google Scholar]
- 52. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64–9. [DOI] [PubMed] [Google Scholar]
- 53. Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res 2013;41:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020;578:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga JI, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015;47:1304–15. [DOI] [PubMed] [Google Scholar]
- 56. Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 2016;534:402–6. [DOI] [PubMed] [Google Scholar]
- 57. Yoshizato T, Nannya Y, Atsuta Y, Shiozawa Y, Iijima-Yamashita Y, Yoshida K, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood 2017;129:2347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shiozawa Y, Malcovati L, Galli A, Sato-Otsubo A, Kataoka K, Sato Y, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boeva V, Zinovyev A, Bleakley K, Vert JP, Janoueix-Lerosey I, Delattre O, et al. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 2011;27:268–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Townsend EC, Murakami MA, Christodoulou A, Christie AL, Koster J, DeSouza TA, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell 2016;29:574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets of sequencing in samples with AEL are available in the European Genome−phenome Archive database (Accession ID: EGAS00001003696 and EGAS00001005810).



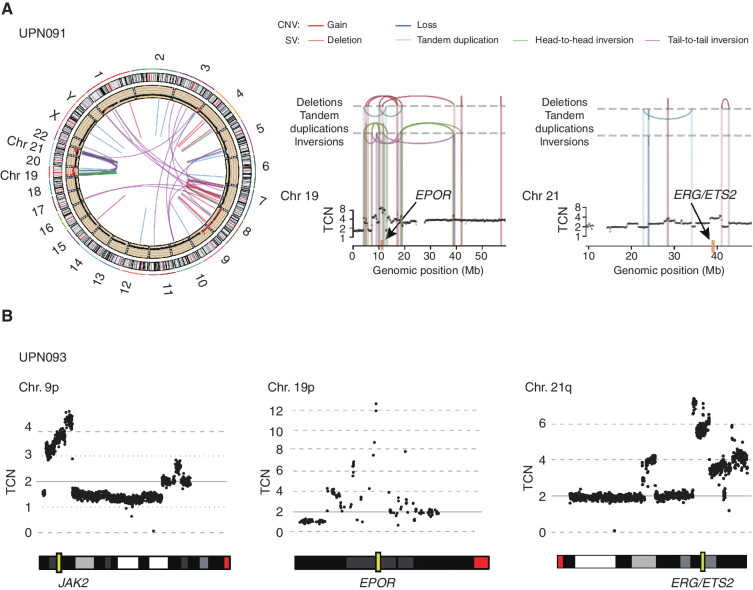
![Figure 2. Global genetic profile in acute erythroleukemia. Genomic landscape of 124 AEL cases. Each column indicates a patient. The heatmap demonstrates mutations, SVs, and CNAs. Genetic groups were clustered according to the status of genetic events. Cases with AEL were divided into four groups based on the correlation of frequent mutations and SVs [A: TP53-mutated AEL, B: NPM1-mutated AEL, C: STAG2-mutated but NPM1-intact AEL, D: other (triple-negative, TN) AEL]. UPD: uniparental disomy.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e773/9894574/dbb80aa854a7/410fig2.jpg)