Extended Data Figure 1: S2G strategy linking each SNP to best gene leads to higher precision than linking SNPs to multiple target genes.
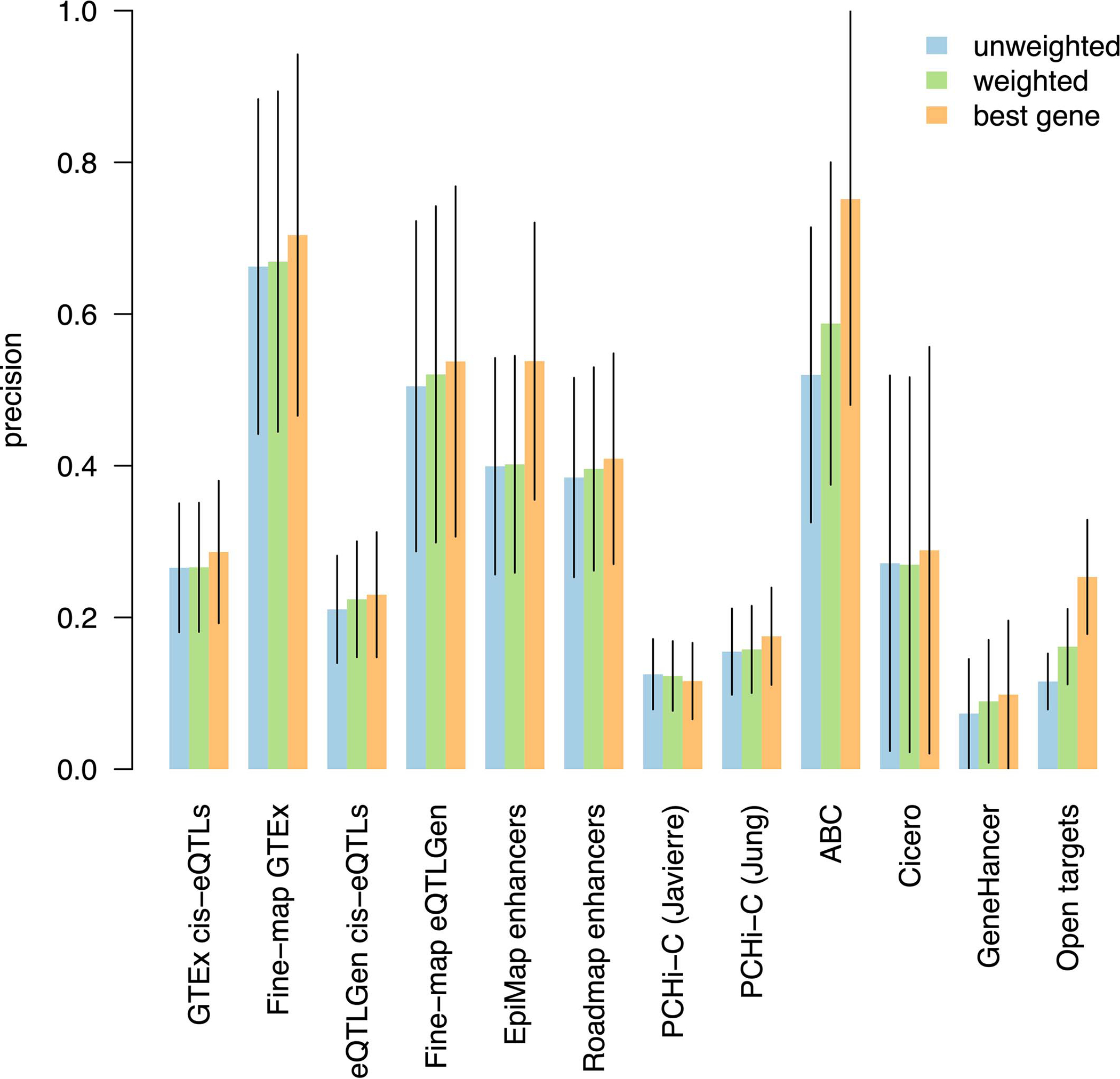
We report the precision of S2G strategies linking SNPs to target genes using three difference approaches for converting raw linking values into linking scores: by assigning to each gene with non-zero raw linking value the same linking score (unweighted), by assigning to each gene a linking score proportional to its raw linking value (weighted), and by retaining only the gene(s) with the highest linking score (best gene). Values were estimated using non-trait-specific training critical gene set and meta-analyzed across 63 independent traits. Error bars represent 95% confidence intervals around meta-analyzed values. For most of the S2G strategies the precision was very similar (except for EpiMap, ABC and Open Targets), but the precision was generally highest for the “best gene” strategy. However, we note that this choice does not reflect biological reality, in which a regulatory element may target more than one gene, and that refinements to this choice are a direction for future research.
