Extended Data Figure 3: Precision of functional S2G strategies using all available cell-types and tissues or restricted to blood and immune cell-types and tissues.
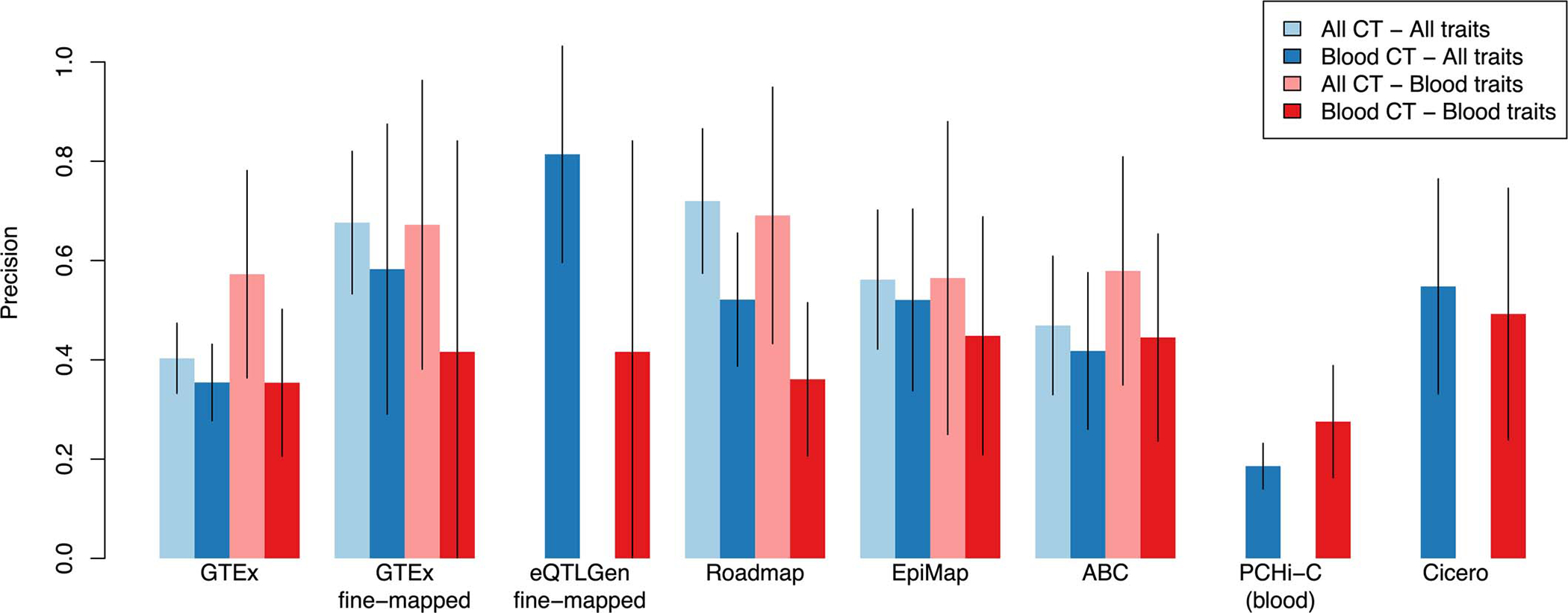
We report the precisions of functional S2G strategies built using either all available cell-types and tissues (All CT; in light color) and/or blood and immune cell-types and tissues (Blood CT; in dark color) meta-analyzed across 63 independent traits (All traits; in blue) and 11 blood cell traits and autoimmune diseases (Blood traits; in red) (UK Biobank all auto-immune diseases, Crohn’s Disease, Rheumatoid Arthritis, Ulcerative Colitis, Lupus, Celiac, Platelet Count, Red Blood Cell Count, Red Blood Cell Distribution Width, Eosinophil Count, White Blood Cell Count; see Supplementary Table 3). Error bars represent 95% confidence intervals around meta-analyzed values. We considered 5 S2G strategies with data available for cell-types and tissues: GTEx cis-eQTLs (GTEx), GTEx fine-mapped cis-eQTL (GTEx fine-mapped), Roadmap enhancer-gene linking (Roadmap), EpiMap enhancer-gene linking (EpiMap), and Activity-By-Contact (ABC). We considered 3 S2G strategies with data available only for blood and immune cell-types and tissues: eQTLGen fine-mapped blood cis-eQTL (eQTLGen fine-mapped), PCHi-C (blood), and Cicero blood/basal (Cicero). We observed 1) that S2G strategies using data from all cell-types and tissues were more precise than S2G strategies restricted to blood and immune cell-types and tissues in both analyses of all traits (light blue vs. dark blue) and blood cell traits and autoimmune diseases (light red vs. dark red), and 2) that S2G strategies using data from blood and immune cell-types and tissues are more precise in all traits than in blood cell traits and autoimmune diseases (dark blue vs. dark red).
