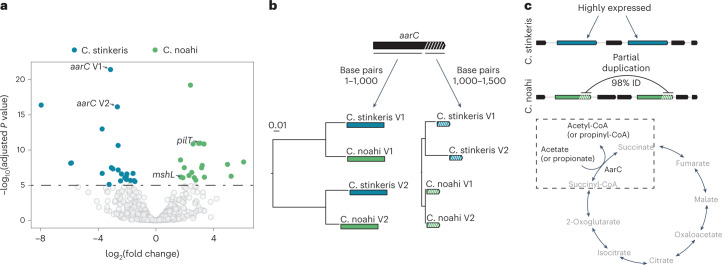
Fig. 4. In vivo expression and analysis of evolution of an acetate acetyl-CoA transferase (AarC) that differentiates ‘Ca. C. stinkeris’ and ‘Ca. C. noahi’.
a, Volcano plot showing the range of fold changes and corresponding adjusted P values from differential expression analysis using DESeq2 comparing the two populations with the metatranscriptomic data from Mann et al.13 (Supplementary Table 1). The horizontal line indicates significant genes with a adjusted P-value cut-off of 0.00001 (n = 6, Benjamini–Hochberg method for adjusted P values, two-sided; Supplementary Table 4). The two variants of aarC are marked. mshL is also marked as it corroborates the analysis in Fig. 3. b, Phylogenetic trees at the gene-level for different regions of the two variants of aarC. The two trees were reconstructed using different segments of the four aarC variants (two per genome), either the first 1,000 bp or the last 500 bp. c, Graphical summary of results and hypothesis from the analysis in a and b. The gene diagrams show that two different variants of the aarC gene are highly expressed in ‘Ca. C. stinkeris’ relative to ‘Ca. C. noahi’ wherein a segment of the aarC genes has been homogenized. AarC is CoA transferase predicted to assimilate acetate via the tricarboxylic acid cycle, but has also been shown to have activity on propionate, leading to propionyl-CoA48.