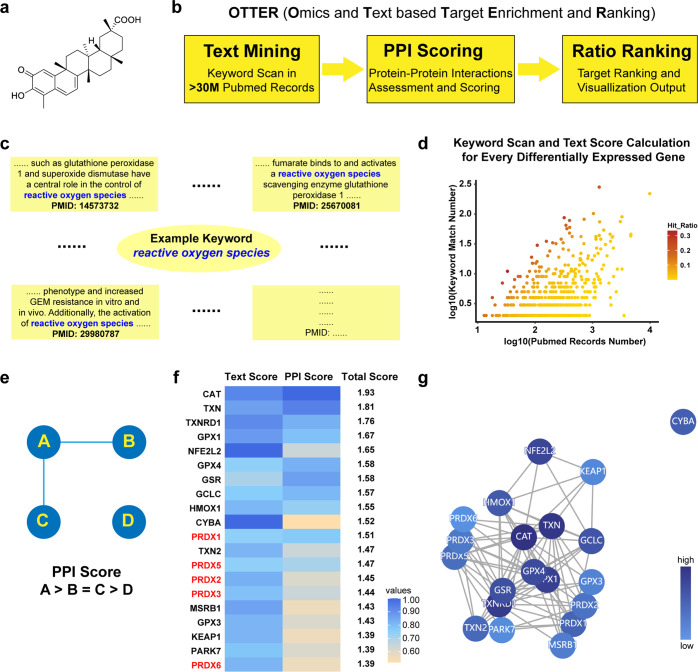
Fig. 1.
A new computational tool named OTTER for target identification. a The chemical structure of Celastrol. b The workflow of OTTER consists of three steps, including text mining in PubMed abstracts, scoring with protein-protein interactions, and ranking with visualization. c Illustrative diagram for the text mining process of OTTER. Several abstracts containing the keyword “reactive oxygen species” were chosen as an example. For each gene in the list provided by users, the user-defined keyword is scanned in all the PubMed abstracts of this gene. d Text mining and scores calculation are performed for every differentially expressed gene in the list provided by users. For each gene, a hit ratio is calculated using the Keyword Match Number divided by the PubMed Record Number. Then a hit ratio is normalized as a Text Score for every gene in the list. e The rule used by OTTER for protein-protein interaction assessment. After the calculation of Text Scores, these genes are scored with the protein-protein interaction records from the STRING database. The genes involved in more protein-protein interactions with other genes in the list are given higher PPI Scores. f The top-ranking 20 differentially expressed genes according to total scores calculated by OTTER, using the RNA sequencing data from cells treated with Celastrol, and the keyword “reactive oxygen species”. g The interactive plot generated by OTTER, using top-ranking 20 differentially expressed genes according to final scores. Colored nodes represent top-ranking genes after enrichment, while lines represent protein-protein interactions between these genes. Nodes with higher scores are colored in darker blue