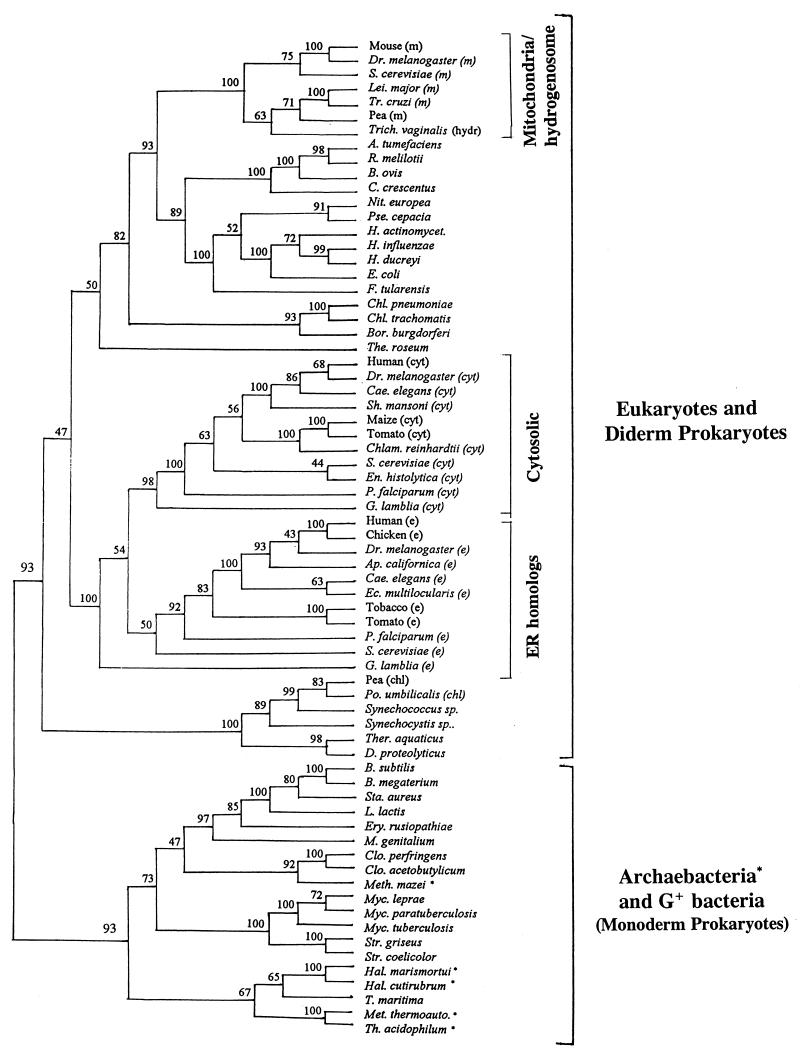
FIG. 27.
A consensus neighbor-joining tree based on Hsp70 sequences (bootstrapped 100 times) showing the relationship between prokaryotic and various eukaryotic homologs. The tree is based on 531 aligned amino acid positions. The main points to be noted are as follows: mitochondrial and chloroplasts homologs show a specific relationship to the α proteobacteria and cyanobacteria, respectively; the hydrogenosome homolog from Trichomonas branches with the mitochondrial clade; the eukaryotic nuclear-cytosolic homologs form a distinct clade within gram-negative bacteria unrelated to the organellar homologs; the ER and cytosolic homologs form paralogous gene families; and archaebacterial homologs (marked with asterisks) show polyphyletic branching within gram-positive bacteria. In the tree shown, only a small number of divergent eukaryotic homologs are included. However, inclusion of additional eukaryotic homologs does not alter the phylogenetic relationship shown here (unpublished results).