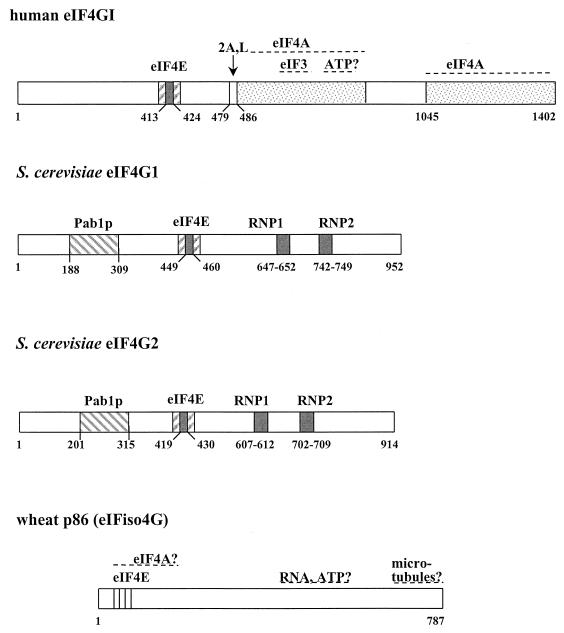
FIG. 5.
This scheme compares the overall structures and known or predicted binding sites of mammalian eIF4GI (242, 312, 346, 605), its two counterparts in S. cerevisiae (180), and wheat p86 (6, 373). The sites of cleavage by the proteases 2A and L and of the RNA-binding motifs (RNP) are also indicated. The potential eIF3 binding and RNA-binding motifs in yeast proteins have been deduced from sequence comparisons. There is no evidence for the existence of an eIF4A-binding site in the yeast eIF4Gs, whereas there are two such domains in mammalian eIF4G (242). The protein structures are approximately arranged in order to line up the homologous regions. Wheat p86 is thought to have a binding site for microtubules at the C terminus (58). There has been uncertainty about the N-terminal sequence of eIF4GI (181), which is now thought to include a Pabp-binding site (509a). Further examples of eIF4G or of eIF4G-like proteins are discussed in the text.