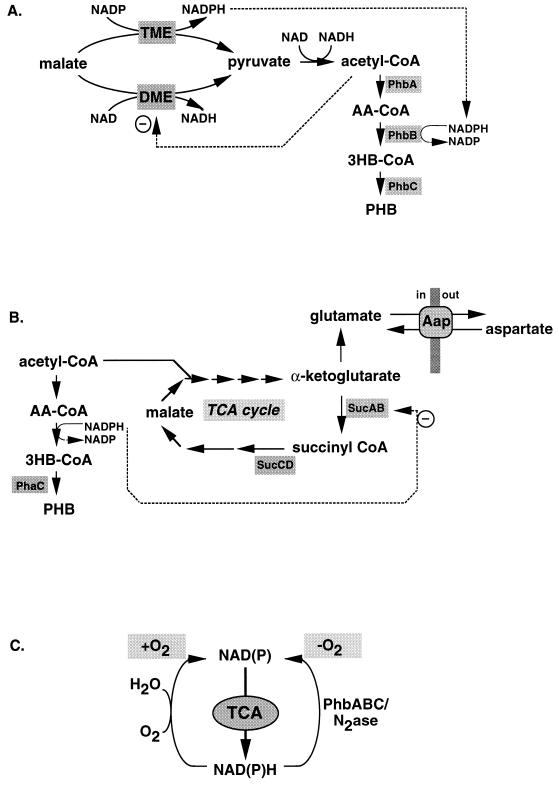
FIG. 11.
P(3HB) metabolism and N2 fixation in Rhizobium. (A) In the bacteroid of R. meliloti in symbiosis with alfalfa, the Tme malic enzyme is not expressed while Dme is inhibited by excess acetyl-CoA. Consequently, the levels of NAD(P)H are too low to pull acetyl-CoA into the P(3HB) pathway. In the free-living state, however, both Tme and Dme are active and P(3HB) formation is initiated under the desired conditions. (B) A direct link in central metabolism between the TCA cycle, P(3HB) formation, and amino acid metabolism is apparent from studies of the R. leguminosarum amino acid permease. Mutants that are less sensitive to high levels of aspartate have an increased secretion of glutamate. This increased production of glutamate is caused by inhibition of the TCA cycle either by a mutation in one of the genes encoding a TCA cycle enzyme or by a mutation in the PHA polymerase gene. In the absence of P(3HB) synthesis, the TCA cycle cannot function optimally, since increased reducing equivalents inhibit α-ketoglutarate dehydrogenase. Both types of mutations cause accumulation of α-ketoglutarate, which is directly converted to glutamate. (C) Recycling of reducing equivalents in Rhizobium. The TCA cycle is the most important pathway for supplying precursors of amino acids. To keep the TCA cycle active in the anaerobic bacteroid, P(3HB) biosynthesis and nitrogenase oxidize reducing equivalents. Different Rhizobium spp. have evolved different means to regulate the three NAD(P)H-oxidizing pathways in the free-living or bacteroid state.