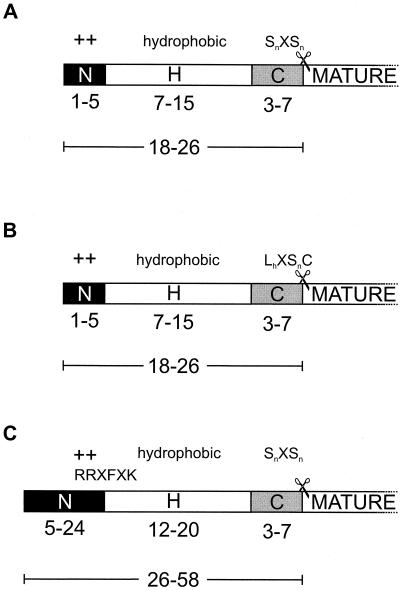
FIG. 1.
Domain structure of the signal sequence of precursor proteins. (A) Signal sequences of SRP- or SecB-dependent preproteins have a net positive charge in the N region (indicated by +), a hydrophobic H region, and a C region with the signal peptidase cleavage site (✂) preceded by the motif SnXSn, in which Sn stands for an amino acid with a small neutral side chain and X stands for any amino acyl residue. (B) Type II signal sequences LhXSnC, in which Lh stands for an amino acid with a large hydrophobic side chain and C stands for cysteine. The cleavage site is located between Sn and C. (C) Signal sequences of precursor proteins that are dependent on the twin-Arg route resemble normal signal sequences but have an extended N region and possess the RRXFXK motif, which straddles the H and C domains (9). For both types of signal sequences, the variation in length of the different regions and of the total signal sequence is indicated.