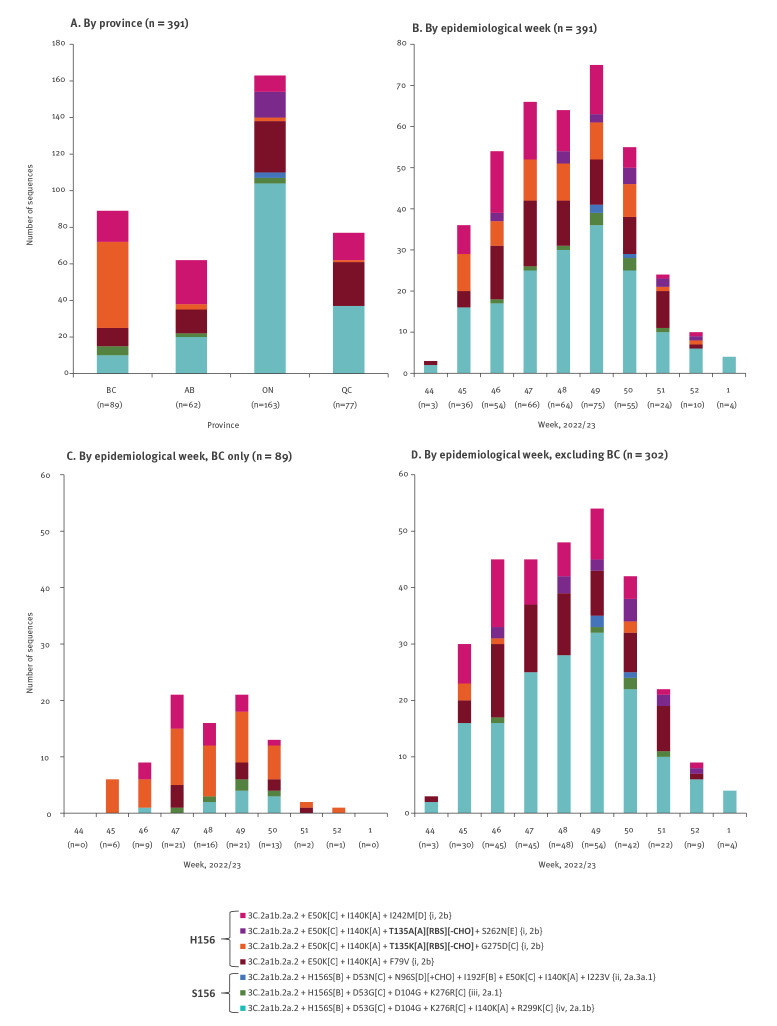
Figure 2.
Distribution of clade 3C.2a1b.2a2 variants among genetically characterised influenza A(H3N2) viruses contributing to influenza vaccine effectiveness analyses, SPSN, Canada, 1 November–28 December 2022 (weeks 44–1) (n = 391)
BC: British Columbia; CHO: carbohydrate; ECDC: European Centre for Disease Prevention and Control; RBS: receptor-binding site; SPSN: Sentinel Practitioner Surveillance Network.
−CHO and +CHO refer to mutations resulting in the loss or gain of a potential glycosylation site, respectively. Genetically characterised influenza A(H3N2) variants contributing to influenza vaccine effectiveness analyses with associated substitutions and ECDC subgroup designations, followed by Nextstrain nomenclature, noted in curly brackets. Colour coding corresponds with subgroups displayed in Supplementary Table S1, the latter including further substitutions and neuraminidase characterisation.