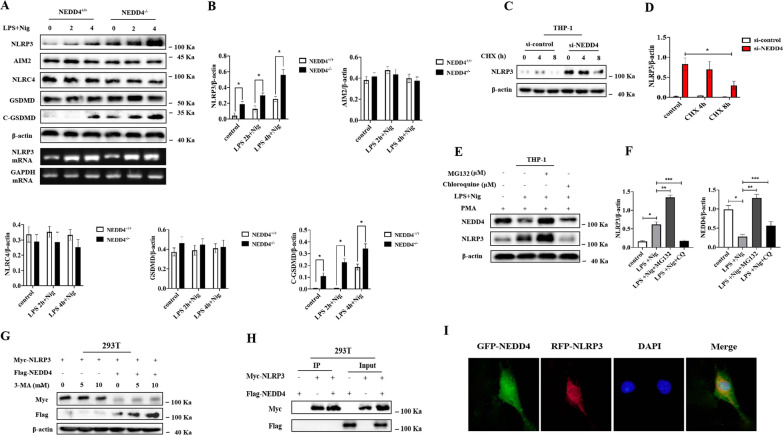
Fig. 5.
NEDD4 indirectly promotes proteasomal degradation of NLRP3. A, B Immunoblot analysis and quantification by densitometry of NLRP3, NLRC4, AIM2, GSDMD and C-GSDMD protein or RT-PCR analysis of mouse peritoneal macrophages from NEDD4+/+ or NEDD4−/− mouse, and then stimulated with LPS/Nig at different times. Data are expressed as mean ± SD (n = 3). *P < 0.05 vs. NEDD4+/+group. C, D Immunoblot analysis and quantification by densitometry of NLRP3 from THP-1 cells silenced of NEDD4, and then treated for various times with cycloheximide (CHX). Data are expressed as mean ± SD (n = 3). *P < 0.05 vs. ctrl siRNA group. E, F Immunoblot analysis of extracts from THP-1 cells stimulated with LPS for 4 h, and followed by stimulation with Nig for 1 h. Then treated with MG-132 and chloroquine (CQ) for 4 h. Data are expressed as mean ± SD (n = 3). *P < 0.05 vs. LPS+Nig, **P < 0.05 vs. LPS+Nig+MG132, ***P < 0.05 vs. LPS+Nig+CQ. G Immunoblot analysis of extracts from HEK293T cells transfected with Myc-NLRP3 and Flag-NEDD4 expression plasmid then treated with 3-MA. H HEK293T cells were transfected with NEDD4 and NLRP3, and cell lysates were immunoprecipitated with anti-Flag or anti-Myc antibody. The immunocomplex was analyzed by immunoblotting. I CMECs transfected with GFP-NEDD4 and RFC-NLRP3 were fixed and incubated with a secondary antibody conjugated to Alexa Fluor 594 and Alexa Fluor 488. Non-colocalization between NEDD4 and NLRP3 was examined by fluorescence microscope