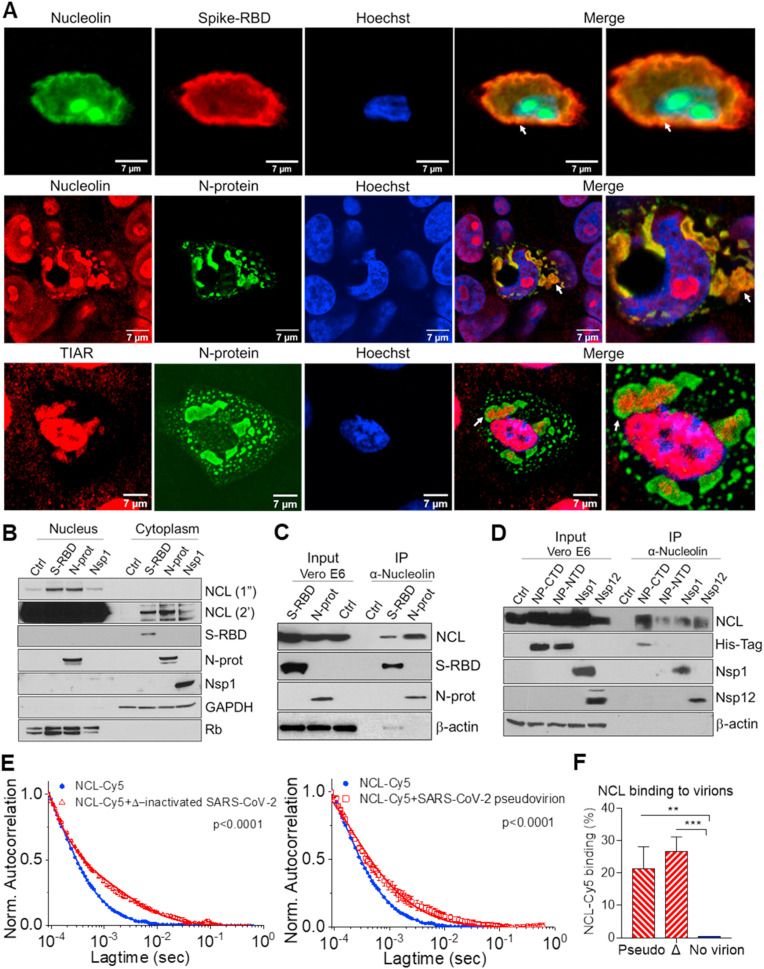
Fig. 1.
SARS-CoV-2 viral proteins interact with nucleolin and induce its cytoplasmic translocation. A. Immunofluorescence staining and confocal images of Vero E6 cells transfected with spike-RBD and N-protein for 48h, with anti-nucleolin (NCL, green or red), anti-spike-RBD (red), N-protein (green), TIAR (red), and Hoechst (blue). The merge of the fluorescent channels, including higher magnification, are shown (right). White arrows indicate co-localization. B. Western blot determination of NCL and SARS-CoV-2 proteins in the nuclear and cytoplasmic fractions of Vero E6 cells transfected with control (ctrl) empty plasmid (pcDNA3.1+), spike-RBD, N-protein and Nsp1 for 48h. Rb and GAPDH were used as nuclear and cytoplasmic markers, respectively. Exposure time for NCL development were 1 s (1″) and 2 min (2′). Lysates from Vero E6 cells transfected with spike-RBD, total N-protein, and control (pcDNA3.1+) (C) and C- and N-terminal domains (conjugated to a histidine-tag), Nsp1 and Nsp12/RdRp (D) were co-immunoprecipitated with NCL. Western blot determination of NCL and SARS-CoV-2 proteins, using total protein lysate (input) or immunoprecipitated complexes (IP). β-actin: loading control. Fluorescence correlation spectroscopy (FCS) auto-correlation plot for Cy5 labeled NCL alone or in complex with SARS-CoV-2 heat-inactivated virion (E) and percentage of NCL-Cy5 binding to virion (F). Average values from triplicate measurements are shown. Error bars indicate standard deviations. *P < 0.05, **P < 0.01 and ***P < 0.001.