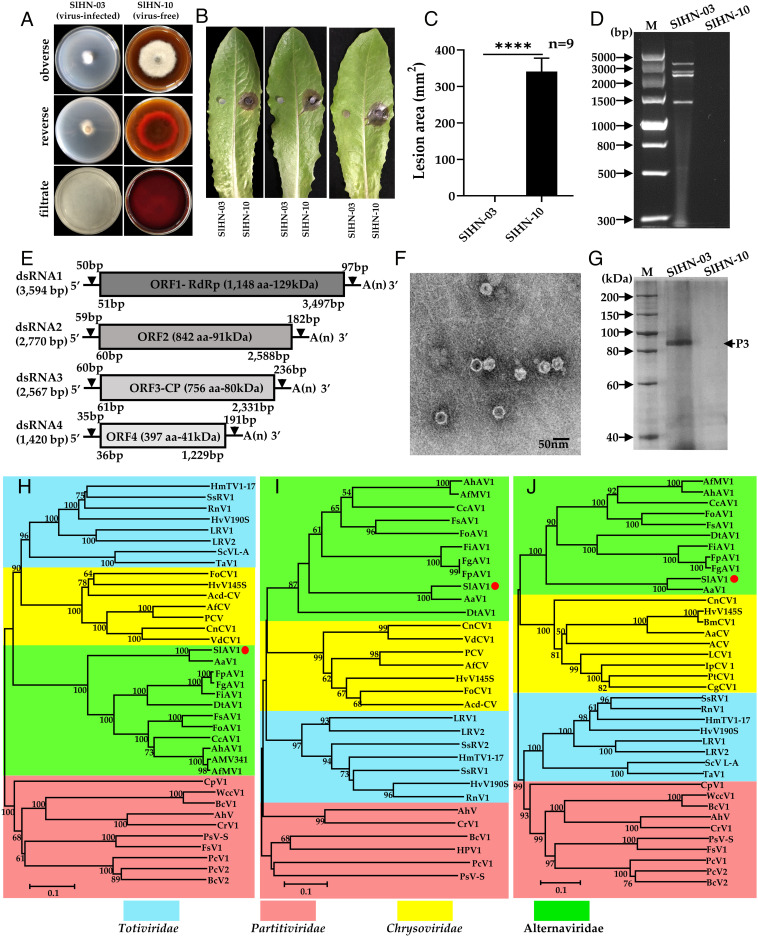
Fig. 1.
Hypovirulence-associated traits of strain SlHN-03 of S. lycopersici and characterization of Stemphylium lycopersici alternavirus 1 (SlAV1). (A) Abnormal colony morphology of virus-free strain SlHN-10 vs virus-infected strain SlHN-03 grown on a PDA plate at 28°C for 7 d. (B) and (C) Virulence assay on detached lettuce leaves (SlHN-10 on the Right and SlHN-03 on the Left). Data are represented as mean ± SD from three replicates with ****P < 0.0001 indicating the statistically significant difference by two-tailed Student’s t test. (D) dsRNA extracted from mycelia of strains SlHN-03 and SlHN-10, lane M: DNA marker (DL5000 bp, Vazyme). (E) Schematic genome organization of SlAV1, gray boxed indicates the putative ORFs encoded by dsRNA1-4, and UTRs as black lines. (F) Electron micrograph of viral particles. (G) Protein components of viral particles were analyzed by SDS-PAGE (8% agar gel); Lane M, protein marker (200 kDa ladder); Neighbor-joining phylogenetic trees constructed based on the amino acid sequences of viral RdRp (H), ORF2 (I) and CP (J) of SlAV1 and selected mycoviruses in the Totiviridae, Chrysoviridae, Partitiviridae, and Alternaviridae families. RdRp, RNA-dependent RNA polymerase. CP, coat protein. The numbers at nodes represent bootstrap values as percentages estimated by 1,000 replicates. The scale bar represents a genetic distance of 0.1 amino acid substitutions per site.