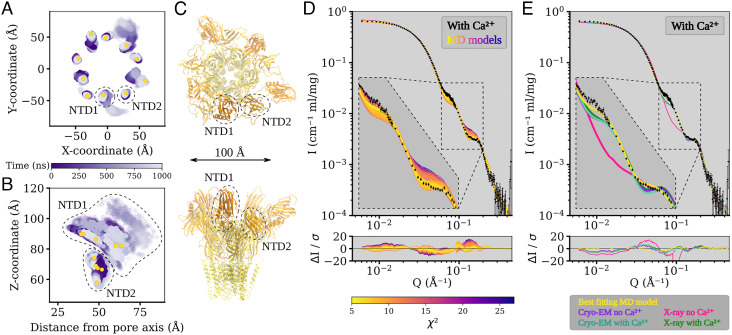
Fig. 5.
The N-terminal domains of DeCLIC are highly dynamic, and an asymmetric arrangement thereof with a mix of contracted and extended NTD positions yields the best fit to the SANS data. (A and B) Position of the center of mass of each NTD lobe over time in four MD simulations, showing their positions in the XY-plane (A) and their Z-coordinate as a function of distance from the pore axis (B). NTD1 is highly mobile, sampling positions further out, lower down, and higher up than the starting conformation. NTD2 has less mobility, mainly sampling positions along Z. Yellow dots show the position of the NTD lobes in C. (C) Snapshot of the MD simulation frame yielding the best fit to the with-calcium SANS data (χ2 of 5.2), seen from the extracellular side (Top) and from the plane of the membrane (Bottom). The protein has adopted an asymmetric conformation with three NTD1 domains in positions similar to the determined structures and two NTD1 lobes further out and down. The snapshot is from the simulations started from the no-Ca2+ cryo-EM structure. (D) Fits of snapshots from simulations of the no-Ca2+ cryo-EM structure and the error-weighted residual between the models and the scattering profile. The simulations yield theoretical distributions around the experimental scattering curve, containing models with better fit (best χ2 of 5.2) to the data than the experimental structure from which the simulations were launched (χ2 of 11.2). Inset shows Q ∈ [0.06, 0.2] Å−1. (E) Comparison between the best-fitting model from simulations (yellow), structures of closed-like DeCLIC (purple, aquamarine, and green; note that curves overlap), and the X-ray no-Ca2+ structure (pink). As seen in the inset showing Q ∈ [0.06, 0.2] Å−1, the simulation model has the best fit to both features in the scattering profile.