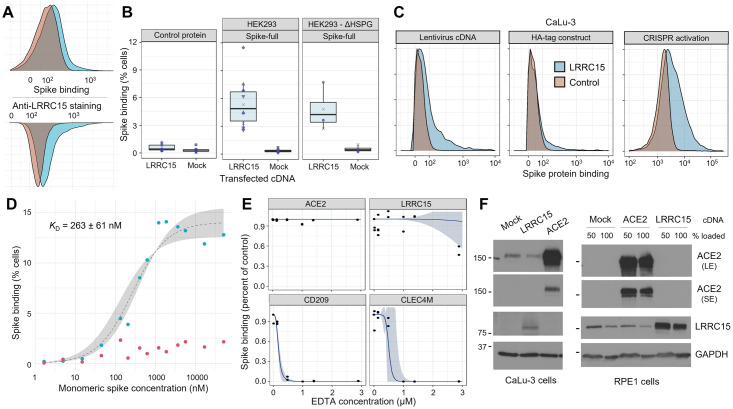
Fig 2. The spike:LRRC15 interaction is robust to cellular context and differs from previously described spike-binding receptors.
(A) Comparison of the cell surface staining profiles of HEK293 cells transfected with LRRC15 cDNA with spike protein (top) and anti-LRRC15 antibody (bottom). (B) LRRC15 binding to spike is specific and independent of heparan sulfate proteoglycans. Boxplots summarizing spike binding of LRRC15-transfected cells compared to mock-transfected cells in wild-type HEK293 cells (center panel), a HEK293 strain deficient in cell surface heparan sulfation following SLC35B2 knockout (right panel) and HEK293 cells binding a control tetramer instead of spike (left panel). (C) LRRC15 expression consistently induces spike binding. LRRC15 overexpression was achieved in a CaLu-3 human lung cell line using three different approaches and increases in spike binding as quantified by flow cytometry. (D) Binding of SARS-CoV-2 spike protein to LRRC15-expressing cells is saturable. Spike binding to LRRC15-expressing (blue) and control (red) cells was quantified by flow cytometry over a wide range of fluorescently conjugated monomeric spike concentrations. A sigmoidal regression curve was fit (gray) to estimate the equilibrium dissociation constant. (E) EDTA blocks known lectin receptors for spike protein but does not prevent LRRC15 binding. Dose–response curves (blue) are fit to flow cytometry measurements of spike binding to HEK293 cells under different concentrations of EDTA. (F) Expression of LRRC15 is not linked to ACE2 protein production. Western blots with antibodies against human ACE2 and LRRC15 detect no up-regulation of ACE2 when LRRC15 is overexpressed or vice versa in either an ACE2-negative cell line (RPE1) or ACE2-positive cell line (CaLu-3). ACE2 is shown as both short (SE) and long (LE) exposures. Molecular masses are indicated in units of kilodaltons. ACE2, angiotensin-converting enzyme 2; LRRC15, leucine-rich repeat containing protein 15; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.