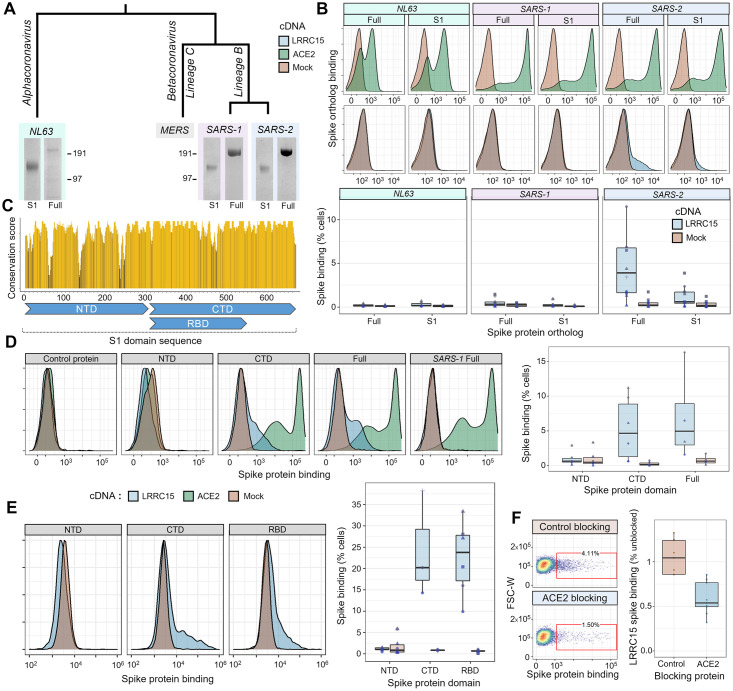
Fig 3. LRRC15 uniquely interacts with the SARS-CoV-2 spike protein and shares a binding interface with ACE2.
(A) Recombinant expression of spike proteins from across the coronavirus family. A labeled phylogenetic tree indicates the relative divergences of coronaviruses above Coomassie-stained gel images of purified recombinant spike proteins. For each virus, the full extracellular domain was produced along with constructs of only the S1 domain. (B) Orthologous spike proteins bind ACE2, but only SARS-CoV-2 strongly binds LRRC15. Flow cytometry traces of spike proteins binding to HEK293 cells overexpressing ACE2 (green, top) or LRRC15 (blue, bottom) compared to control cells (red). Quantified replicates for LRRC15 binding are displayed as boxplots below. (C) Physical conservation of the amino acids in SARS-CoV-2 spike compared to SARS-CoV-1. The rolling average of physicochemical conservation scores across the spike S1 domain sequence is indicated. Regions corresponding to the NTD, CTD, and RBD are annotated below. (D) LRRC15 binding localizes to the spike C-terminal S1 domain. Flow cytometry traces of binding by the CTD but not NTD to LRRC15-transfected cells (left) are shown next to the quantified binding (right). (E) The RBD of the SARS-CoV-2 spike protein is sufficient for binding to LRRC15. The percentage of cells binding each spike truncation construct (right) is shown along with representative flow cytometry traces. These data were collected on a different instrument than those shown in the prior panel, accounting for the variation in scale. (F) Recombinant ACE2 competitively inhibits spike binding to LRRC15. Dotplots of spike binding by flow cytometry to cells where spike protein was preincubated with a control protein or preincubated with the ACE2 extracellular domain. ACE2, angiotensin-converting enzyme 2; CTD, C-terminal domain; LRRC15, leucine-rich repeat containing protein 15; NTD, N-terminal domain; RBD, receptor binding domain; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.