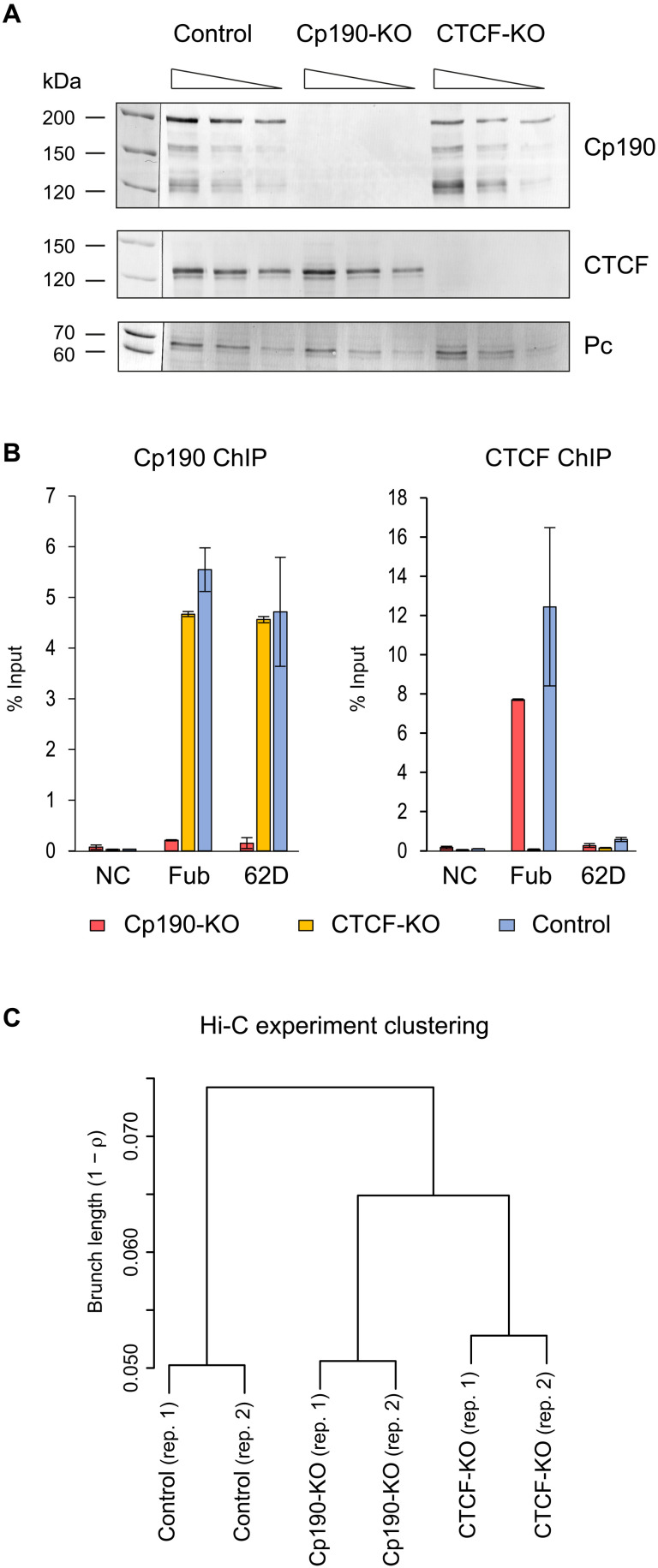
Fig. 1. Characterization of Cp190- and CTCF-deficient cultured cell lines.
(A) Twofold dilutions of total nuclear protein from control (Ras 3), Cp190-KO (CP-R6), and CTCF-KO (CTCF 19.7-1c) cells were analyzed by Western blot with antibodies against Cp190, CTCF, and Polycomb (Pc, loading control). Additional loading control, Coomassie-stained gel of the corresponding total nuclear protein samples, is shown on fig. S2. Positions of molecular weight markers (in kilodaltons) are indicated on the left. (B) ChIP-qPCR demonstrates that Cp190, normally present at Fub and 62D insulators, is no longer detectable at these elements in Cp190-KO cells. Similarly, immunoprecipitation of Fub by anti-CTCF antibodies is abolished by CTCF-KO. Histograms show the average of two independent ChIP-qPCR experiments with whiskers indicating the scatter between individual measurements. An intergenic region from chromosome 3L that does not bind any insulator proteins was used as a negative control (NC). (C) Hierarchical clustering of Hi-C experiments based on pairwise Spearman correlation coefficients (average ρ for the group). To suppress spurious experimental noise, contacts within individual 40-kb bins (the diagonal of contact matrix) and contacts between bins separated by more than 1.6 Mb were excluded from calculations. For bins equal or larger than 40 kb, the clustering is robust to parameter changes (fig. S4).