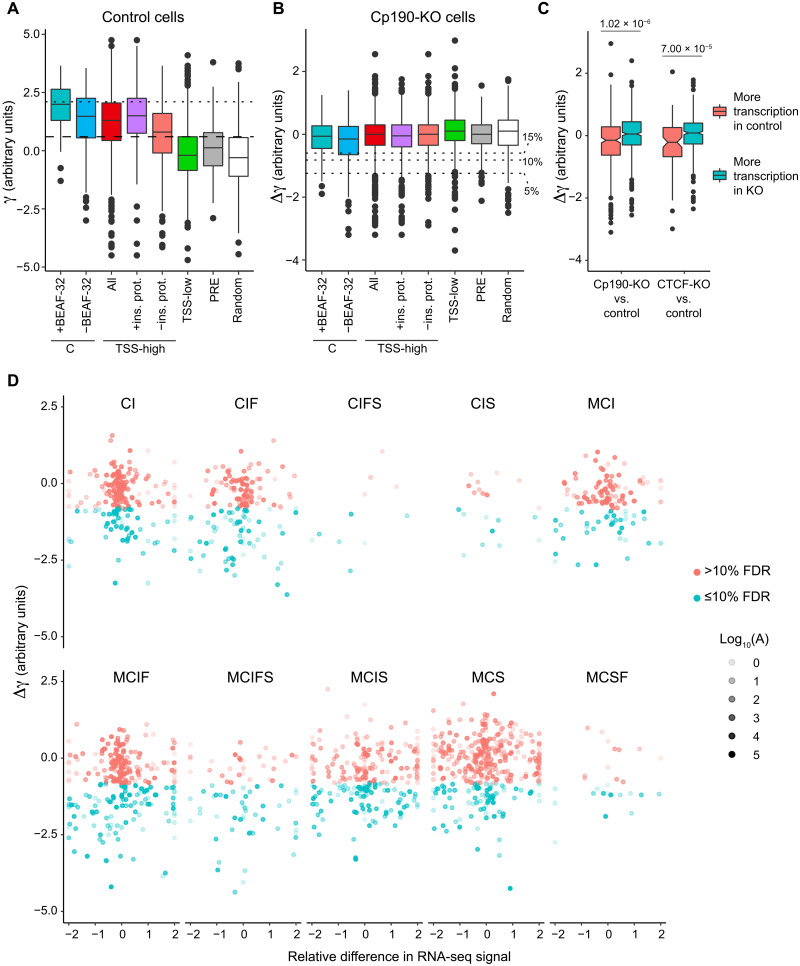
Fig. 5. Transcription start sites of transcriptionally active genes impair chromatin contact crossing.
(A) Box plots display distance-scaling factors (γ) in control cells at class C regions either cobound or not bound by BEAF-32. Also shown are γ values of transcription start site (TSS) grouped by transcriptional activity of the corresponding gene (TSS-high/all and TSS-low) and the binding of insulator proteins mapped in this study (TSS-high/+ins. prot and TSS-high/−ins. prot). Distance-scaling factors of PREs and randomly selected regions with no appreciable ChIP-seq signal for any of the insulator proteins are shown for comparison. Sites with γ above the top quartile in the random control group (horizontal dashed line) are considered as limiting chromatin contact crossing. Horizontal dotted line indicates the top 5% value for γ genome-wide. (B) Cp190 KO leads to no systematic change in distance-scaling factors (Δγ = γKO − γcontrol) at any of the classes of regions. Horizontal dotted lines indicate FDRs used to define insulator elements in Fig. 4. (C) Box plots of Δγ for TSSs of genes whose transcription differs (|log2 fold change| > 2) between Cp190-KO or CTCF-KO and control cells. Note the significant difference between Δγ for TSS with higher transcription in the mutant cells compared to that of the genes with higher transcription in the control cells (P values from Wilcoxon rank sum test are shown above). (D) Scatterplots compare the changes in distance-scaling factors (Δγ ≤ 10% FDR colored in cyan; Δγ > 10% FDR colored in pink) at insulator protein binding sites of various classes to relative differences in RNA-seq signals of genes closest to these sites after Cp190 KO. The average RNA-seq signals between control and Cp190-KO cells (A) in log10 scale are indicated by variable point intensities.