Figure 1. Leveraging clinical microbiome data sets with computational analyses to identify communities and community members to model in vitro.
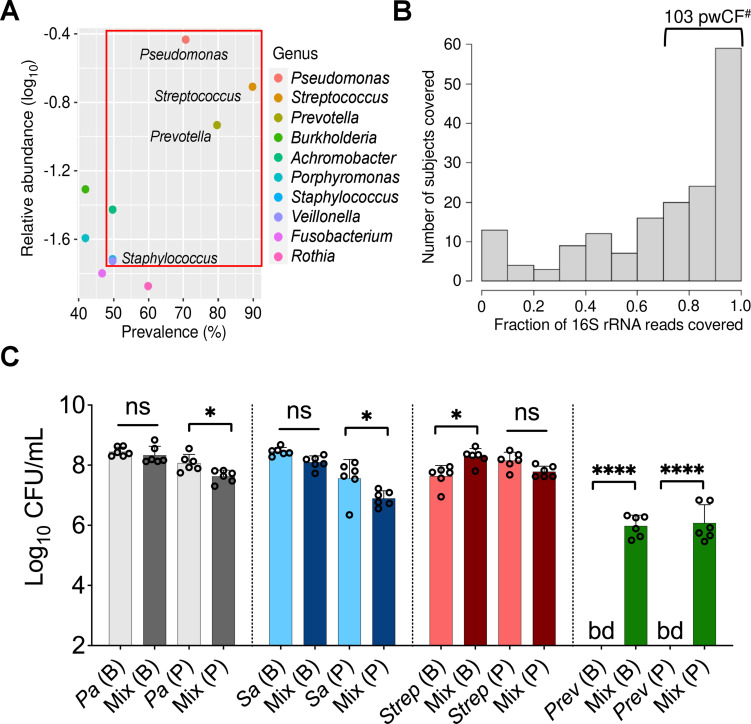
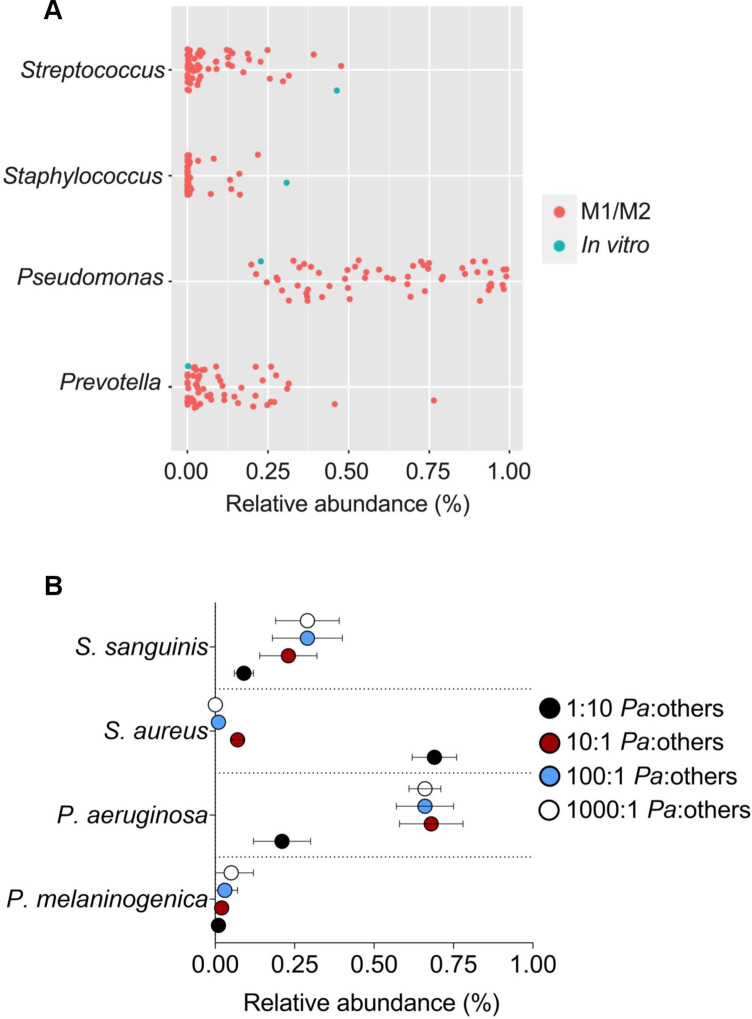
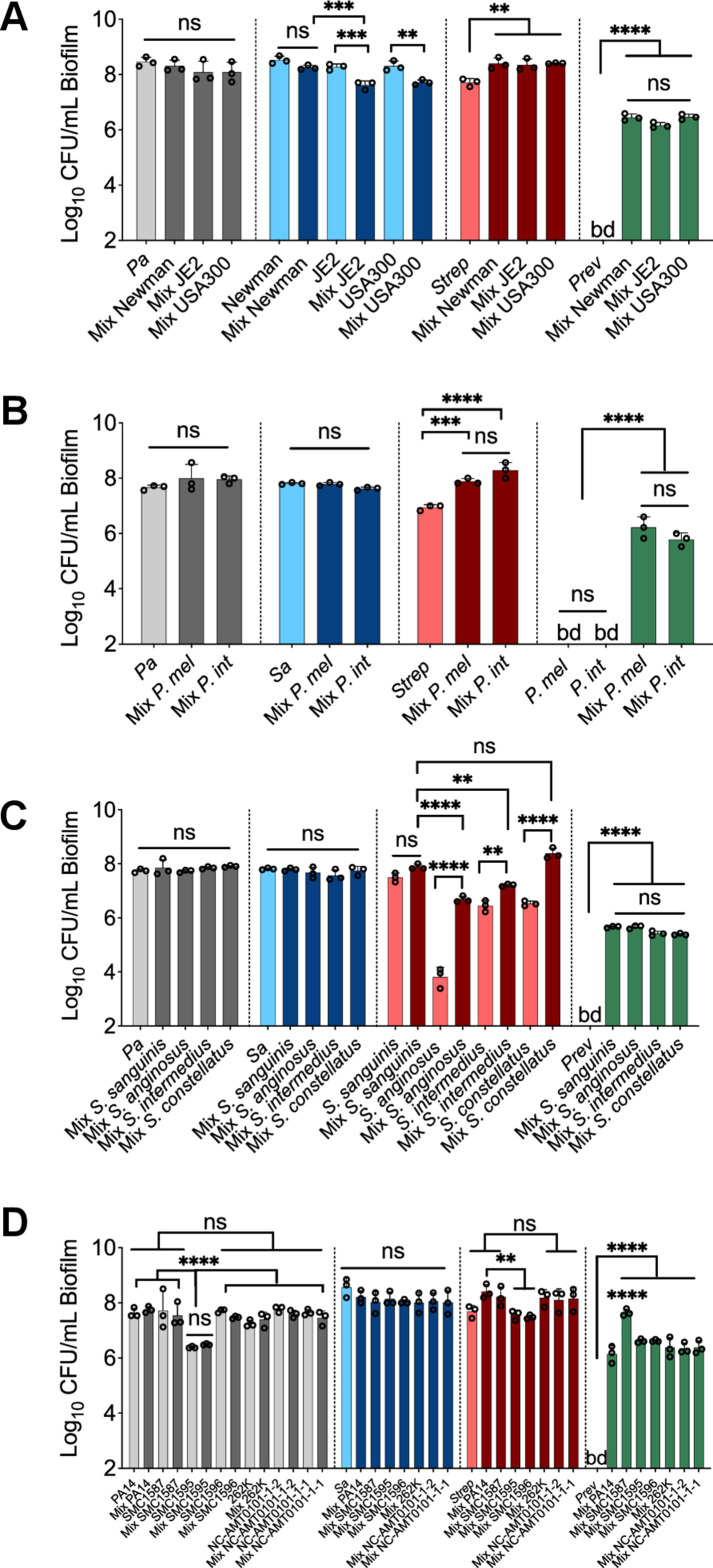
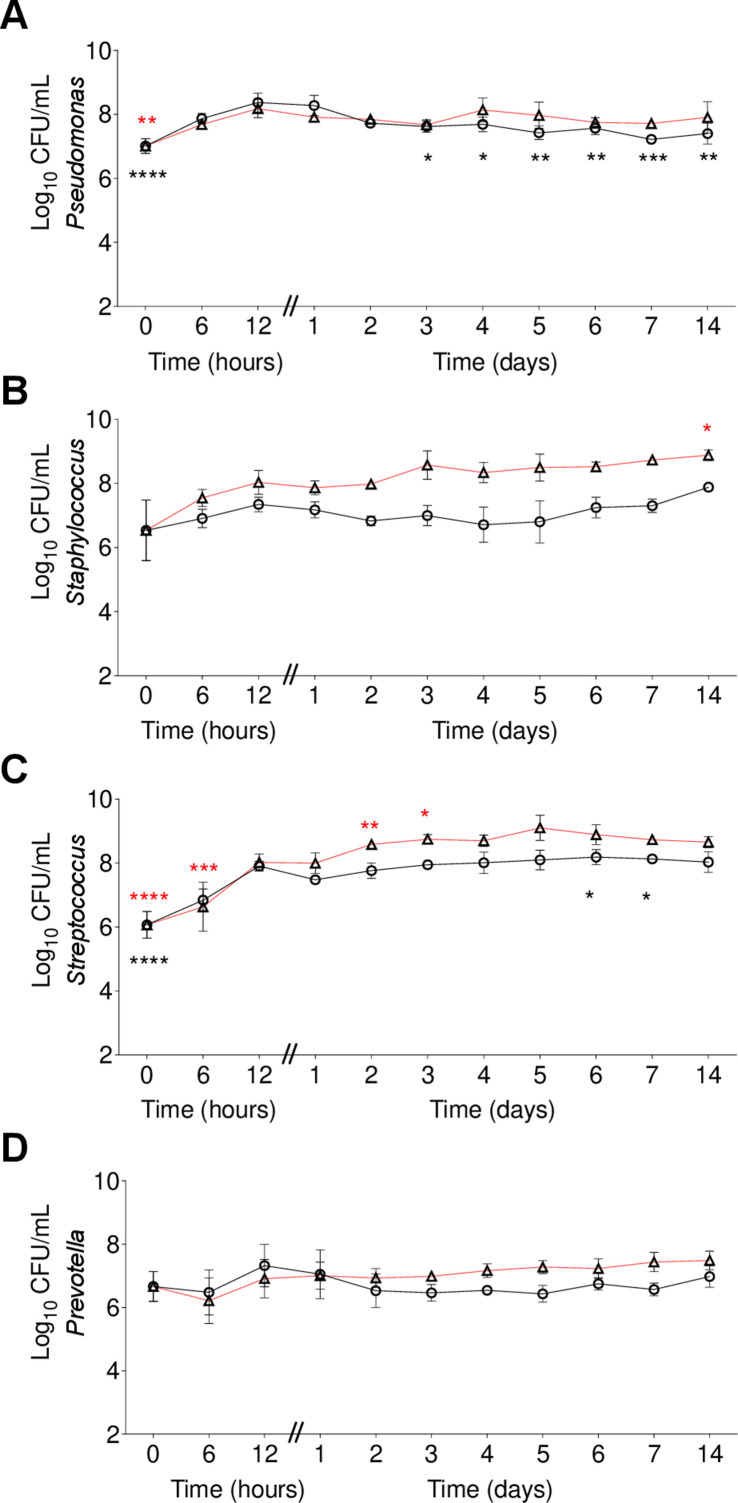
(A) Relative 16S rRNA gene abundance and prevalence of the top 10 cystic fibrosis (CF) lung pathogens in the 167 persons with CF (pwCF) data set used as the basis for developing the in vitro mixed community, as reported by Hampton et al., 2021. (B) Number of unique samples for which >70% of 16S rRNA reads are associated with the combined presence of Pseudomonas, Staphylococcus, Streptococcus, and Prevotella. #Indicates the number of samples (103) that meet this criterion from the total sample size of 167 pwCF. (C) Colony forming units (CFUs) counts of each microbial member grown as a monoculture and in a mixed community (Mix) for biofilm (B) and planktonic (P) fractions. CFUs were performed by plating on medium selective for the growth of each microorganism. Each data point presented in a column represents the average from at least three technical replicates performed at least on three different days (n=6). Statistical analysis was performed using ordinary one-way ANOVA and Tukey’s multiple comparisons posttest with *, p<0.05; ****, p<0.0001, ns = non-significant. Error bars represent SD. Pa = Pseudomonas aeruginosa, Sa = Staphylococcus aureus, Strep = Streptococcus sanguinis, Prev = Prevotella melaninogenica, and bd = below detection.