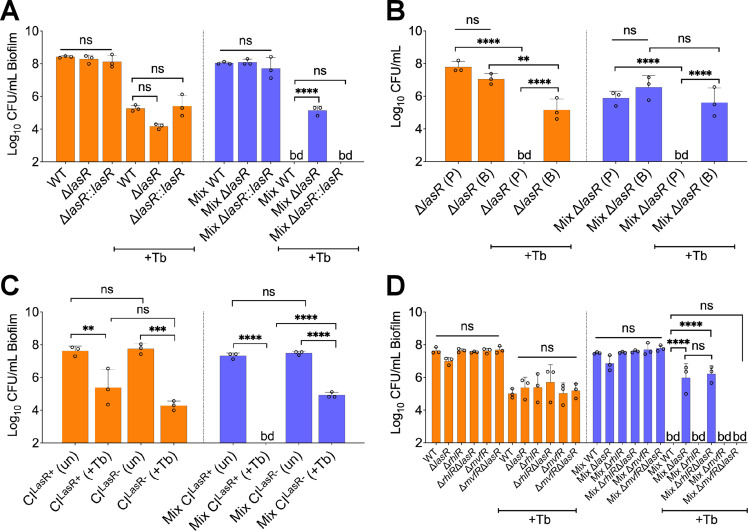
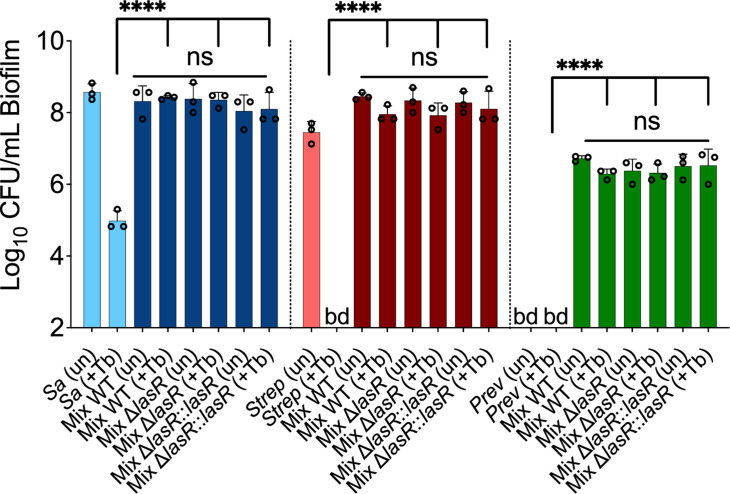
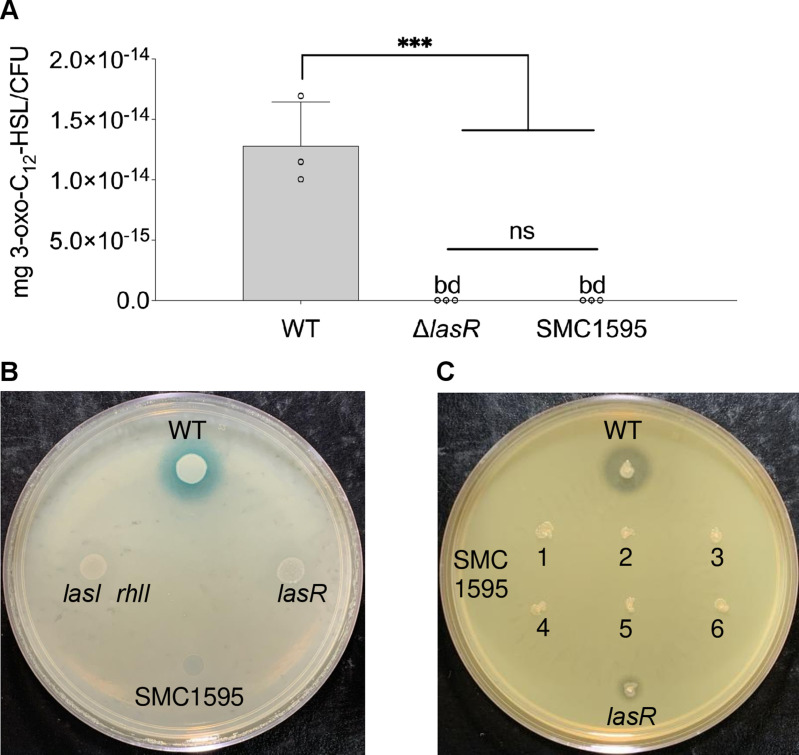
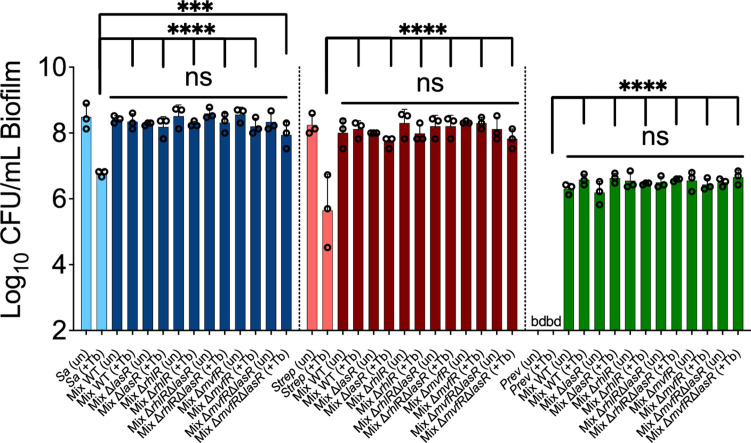
Figure 3. LasR loss-of-function drives biofilm-specific tobramycin tolerance in a mixed community.
Colony forming units of (A) P. aeruginosa strain PA14 (wild-type [WT]), isogenic ΔlasR mutant, and the complemented strain (ΔlasR::lasR), (B) planktonic and biofilm ΔlasR mutant cells, (C) LasR-defective (NC-AMT0101-1-1; LasR−) and LasR+ (NC-AMT0101-1-2) clinical isolates (CIs) and (D) P. aeruginosa quorum sensing regulator mutants grown as monocultures and mixed communities (Mix) and challenged or not with 100 µg/mL of tobramycin (+Tb). Each data point presented in a column represents the average from at least three technical replicates performed at least on three different days (n=3). Statistical analysis was done using ordinary one-way ANOVA and Tukey’s multiple comparisons posttest with **, p<0.01; ***, p<0.001; ****, p<0.0001. Error bars represent SD. bd = below detection and un = untreated.