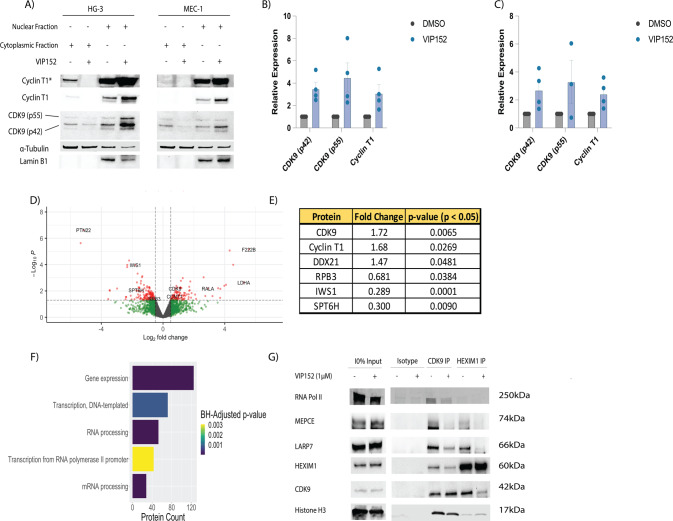
Fig. 4. VIP152 disrupts P-TEFb canonical binding partners.
A–C Cytoplasmic and nuclear extracts of HG-3 and MEC-1 treated with 1 µM VIP152 for two hours with quantification (B, C). D, E Proteomic characterization of nuclear immunoprecipitation of VIP152 treated HG-3 cells. Gray dots indicate non-differentially associated proteins. Green dots indicate transcripts with a LFC > 0.5 or LFC < −0.5 but p-value > 0.05. Blue dots indicate proteins with a p-value < 0.05 but −0.5 < LFC < 0.5. Red dots indicate proteins with a p-value < 0.05 and a LFC > 0.5 or LFC < −0.5. D Volcano plot of differentially associated proteins. E Table of P-TEFb and POLII-associated proteins. F Plot of pathways from Ingenuity Pathway Analysis with p-value < 0.05 and z-score > 0 or z-score < 0 (G) Nuclear IP of CDK9 and HEXIM1 of VIP152 treated HG-3 cells probing for components of the 7SK-RNA complex.