Figure. 2.
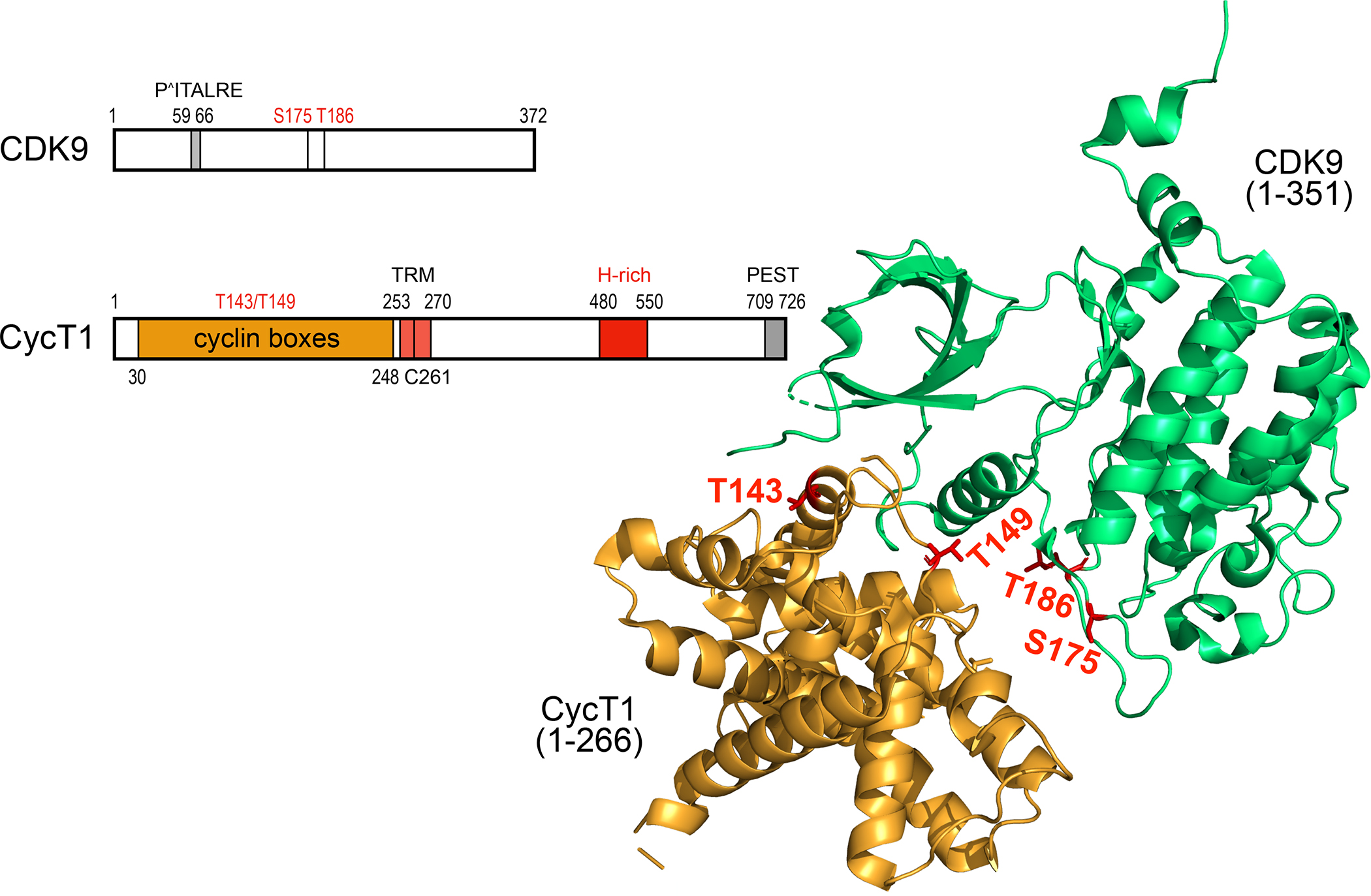
Schematic representation and the 3-D structure of human CDK9 and CycT1 proteins. CDK9 contains 372 residues. From N- to C-termini, important elements contain the PITALRE sequence. serine at positions 175 (S175) and threonine at position 186 (T186, T-loop). S175 and T186 are phosphorylated in the active P-TEFb. CycT1 contains 726 residues. From the N-terminus, there are 2 cyclin boxes (yellow), the Tat-TAR recognition motif (TRM), the histidine-(H)-rich region (red) and the putative PEST sequence (gray). Threonines at positions 143 and 149 (T143. T149) are targets of PKC and facilitate the binding between CDK9 and CycT1. In the unbound CycT1, the degron, where Siah1/2 bind, is located between position 210 and 260. The TRM is the binding site for the HIV transcriptional transactivator Tat. The H-rich sequence binds to targets of CDK9, i.e. the CTD of RNAPII, DSIF and NELF, among others. The function of the PEST sequence remains enigmatic. Some of these features are presented on the 3-D structure of P-TEFb 45,46. The ordered structure (ribbons) contains 266 residues from CycT1 and 351 residues from CDK9. T143 and T149 in CycT1 form intra- and inter-molecular contacts with CycT1 and CDK9, respectively. T186 (T-loop) and S175 in CDK9 are phosphorylated in the active P-TEFb.
