Figure 5.
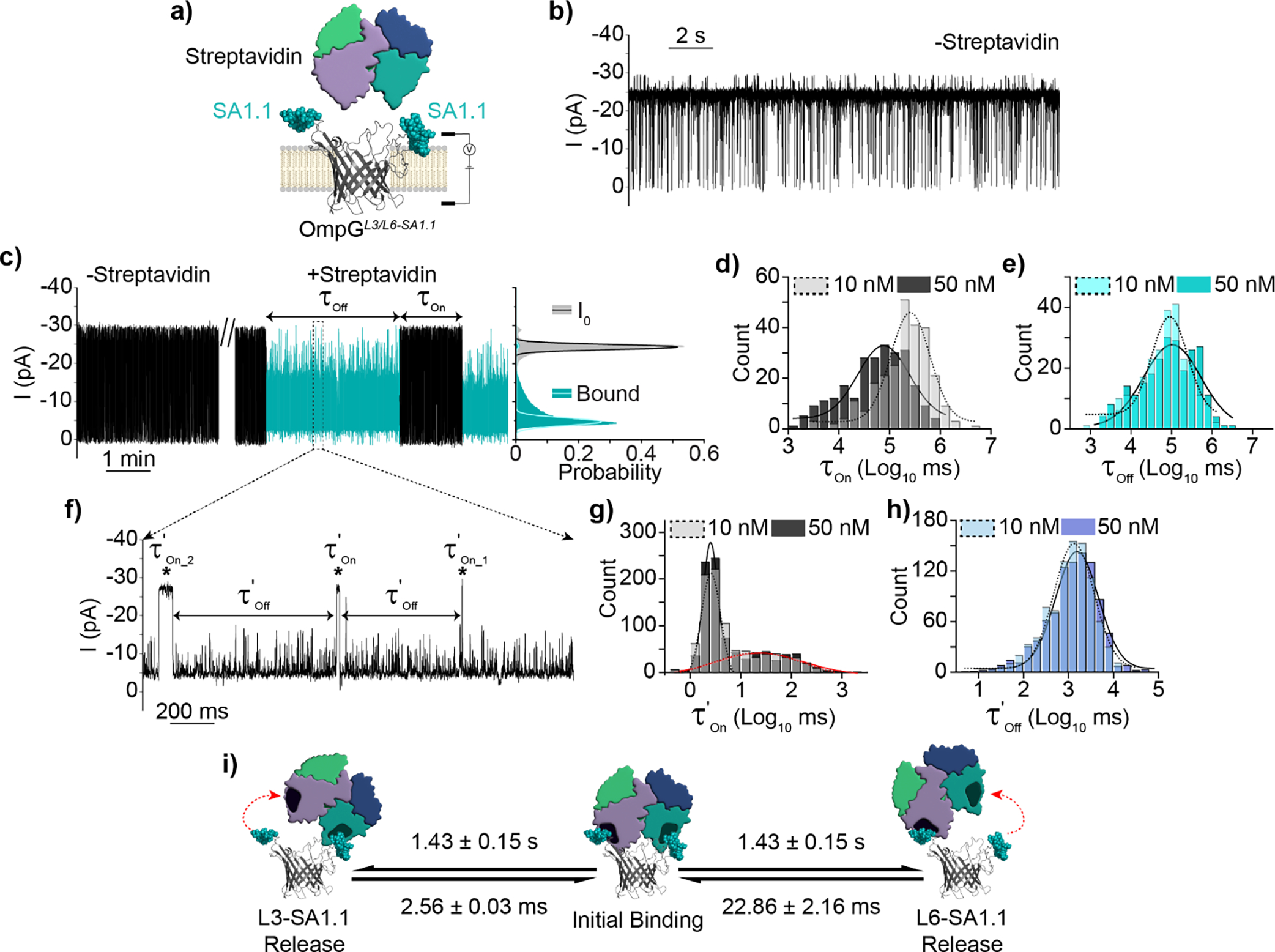
Detection of streptavidin by OmpGL3/L6-SA1.1 avidity nanopore sensor. a) Schematic view of OmpGL3/L6-SA1.1 avidity sensor. b) Representative current trace in the absence of streptavidin c) Representative current trace and all-points histogram in the presence of streptavidin, discrete binding signal(s) indicated in blue. All-points histogram generated from 30 second segments of indicated state. d-e) Gaussian fit of τOn and τOff values derived from discrete streptavidin binding events at either 10 nM (N = 7, n = 258) or 50 nM (N = 5, n = 247). f) Zoomed in view of discrete bound state highlighting streptavidin rebinding sub-states within the discrete signal. g-h) Gaussian fit of kinetic parameters τ’On_1&2 and τ’Off of rebinding events at either 10 nM (N = 3, n = 902) or 50 nM (N = 3, n = 896). i) Proposed model of streptavidin interaction with OmpGL3/L6-SA1.1. Experiments were performed in 50 mM Na2HPO4 pH 6.0 buffer containing 300 mM KCl at an applied potential of ±50 mV. Traces were filtered using a 500 Hz lowpass digital gaussian filter.
