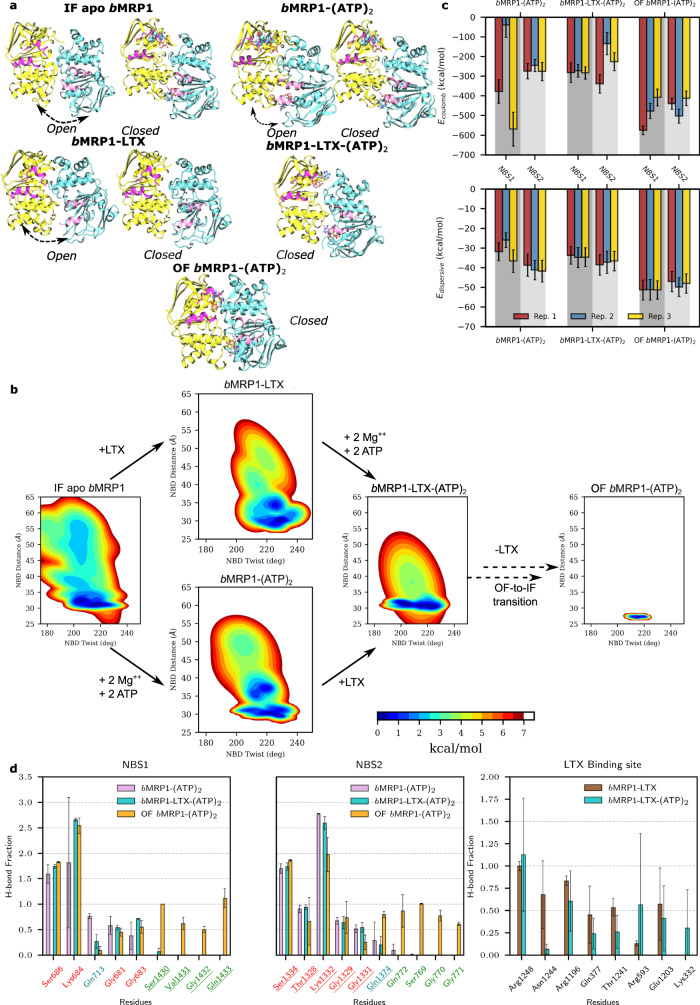
Fig. 2. Asymmetric structural dynamics of bMRP1 systems in POPC:POPE:Chol (2:1:1) and their conformational landscape.
a Representative snapshots picturing the open and closed conformations of NBD dimers observed during MD simulations. IF apo bMRP1, bMRP1-(ATP)2 and bMRP1-LTX revealed two main subpopulations for which black arrows highlight the motion; while the pre- and post-translocation conformations (namely, bMRP1-LTX-(ATP)2 and OF bMRP1-(ATP)2) exhibited only the closed NBD dimer conformation. NBD1, NBD2 and coupling helices are respectively depicted in yellow, cyan and pink. b System-dependent local conformational landscapes obtained from the GMM-based approach developed in the InfleCS method23,64 highlighting the influence of nucleotides and the substrate on bMRP1 structural dynamics. c Averaged Coulomb and van der Waals potentials calculated between nucleotides and NBS1 and NBS2, separately (800 points were used for each replica, treated independently; error bars show standard deviations). d Calculated H-bond networks between ATP and NBS1 or NBS2 and between LTX and substrate-binding pocket residues (H-bond fractions were calculated from each replica independently and were then averaged n = 3; error bars showing standard deviation over them).