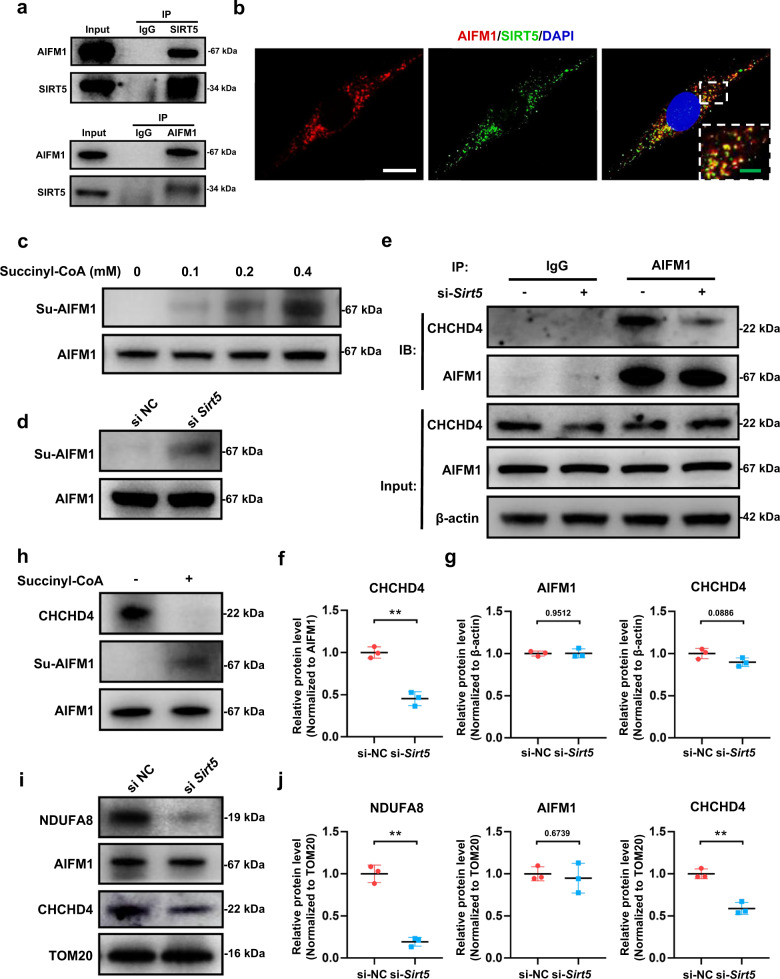
Fig. 5. SIRT5 directly binds to AIFM1 and regulates the succinylation modification of AIFM1 through its desuccinylase activity, thereby regulating AIFM1-CHCHD4 complex function.
The binding of SIRT5 and AIFM1 was verified in rat NP cells. a Co-IP of AIFM1 by SIRT5 antibody and reverse Co-IP of SIRT5 by AIFM1 antibody. b Representative immunohistochemical staining of the intracellular location and distribution of AIFM1 and SIRT5. White scale bar = 20 μm; green scale bar = 5 μm. In the in vitro succinylation assay, AIFM1 was purified from NP cells and incubated with the indicated concentrations of succinyl-CoA in vitro. c Western blotting analysis of succinylated AIFM1 (n = 3). d Western blotting analysis of the succinylation of AIFM1 from rat NP cells in the si-NC and si-Sirt5 groups (n = 3). The binding of CHCHD4 and AIFM1 was detected in rat NP cells from the si-NC and si-Sirt5 groups. e Co-IP of CHCHD4 by the AIFM1 antibody. g Quantification of CHCHD4 and AIFM1 protein expression (normalized to β-actin expression, n = 3). f Quantification of CHCHD4 bound to AIFM1 (normalized to AIFM1 expression, n = 3). An in vitro succinylation assay was performed to verify the disruptive effect of succinylation modification on the binding between AIFM1 and CHCHD4. h Western blotting analysis of succinylated AIFM1 and CHCHD4 protein bound to AIFM1 (n = 3). Mitochondrial proteins were extracted from rat NP cells in the si-NC and si-Sirt5 groups. i, j Western blotting analysis and quantification of NDUFA8, AIFM1, and CHCHD4 protein expression extracted from mitochondria (normalized to TOM20 expression, n = 3). Differences between two groups were analyzed by Student’s t test. The data in the figures represent the mean ± S.D. The P value is shown, **P < 0.01.