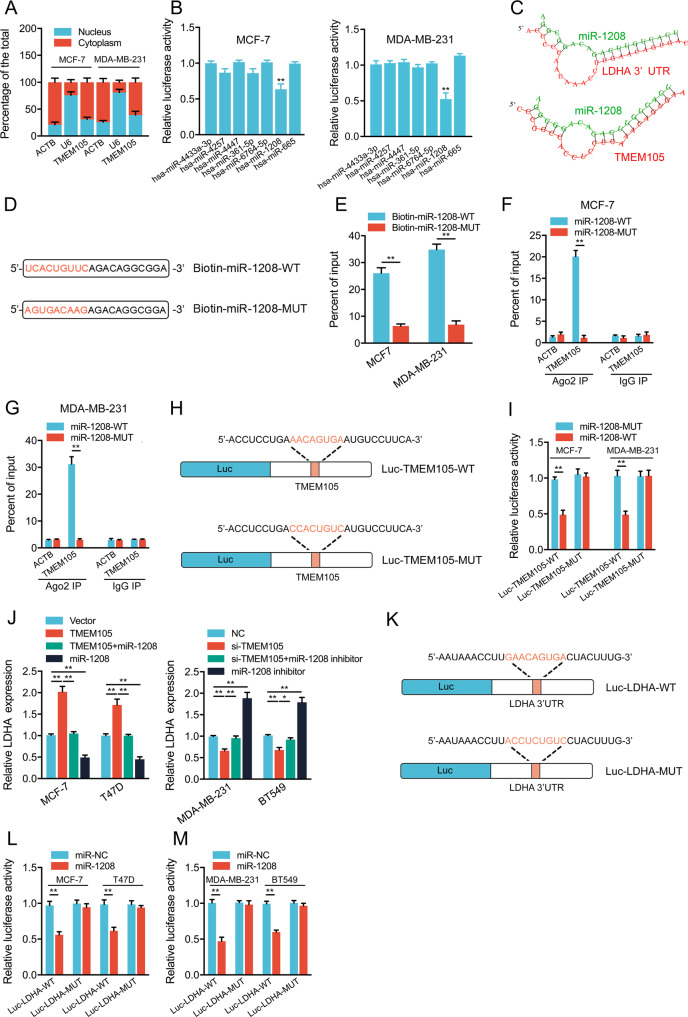
Fig. 5. TMEM105 sponges miR-1208 to upregulate LDHA expression in breast cancer cells.
A The subcellular localization analysis of TMEM105 in breast cancer cells was measured by qRT-PCR. ACTB was the positive control for the cytoplasm, and U6 was the positive control for the nucleus. B Overexpression of the potential candidate miRNAs on the luciferase activity of TMEM105-WT in MCF-7 and MDA-MB-231 cells. C The binding sites in miR-1208, TMEM105, and the LDHA 3′UTR were predicted using the RNAhybrid program. D The schematic images of miR-1208-WT and miR-1208-MUT. E MCF-7 and MDA-MB-231 cell lysates were incubated with biotin-labeled miR-1208-WT or miR-1208-MUT. The expression of TMEM105 in pull-down products was analyzed by qRT-PCR. F, G Ago2-RIP followed by qRT-PCR to evaluate TMEM105 expression after transfection with miR-1208-WT or miR-1208-MUT in MCF-7 (F) and MDA-MB-231 (G) cells. H The schematic images of reporter genes contained TMEM105-WT or TMEM105-MUT without the miR-1208 binding site. I Effects of miR-1208-WT or miR-1208-MUT on the luciferase activity with the TMEM105-WT or TMEM105-MUT in MCF-7 and MDA-MB-231 cells. J Effects of miR-1208 mimics or inhibitors on LDHA levels in the TMEM105-overexpressing or knockdown cells. K The schematic images of reporter genes contained LDHA 3′ UTR wild or the mutant type without the miR-1208 binding site. (L, M) Effects of miR-1208 on the luciferase activity with the LDHA-WT or LDHA-MUT in breast cancer cells. *p < 0.05, **p < 0.01.