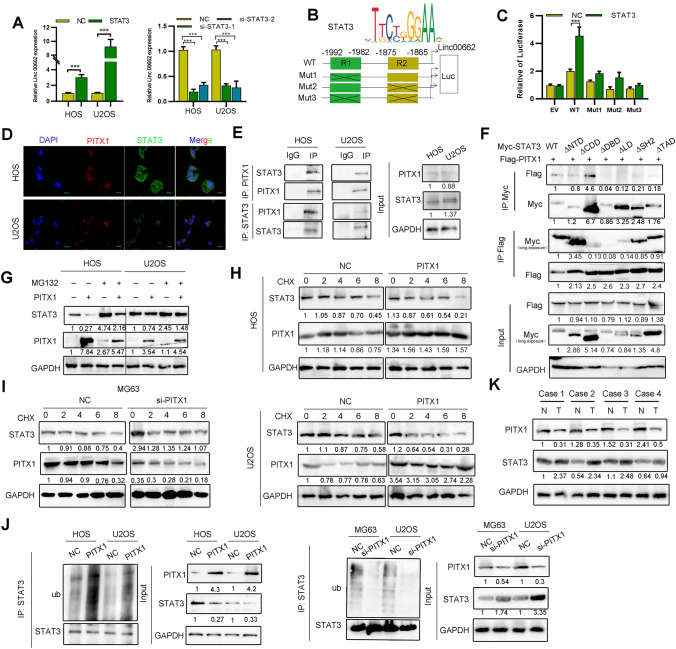
Fig. 4.
PITX1 promotes STAT3 proteasome degradation to inhibit LINC00662 expression. A Expression of LINC00662 was detected after STAT3 overexpression or knockdown in OS cells. B Online JASPAR software was used to predict the possible binding sites (R1 and R2) of STAT3 in the LINC00662 promoter. Schematic diagram of luciferase reporter vectors containing wild-type (WT) or mutant (Mut1: R2 mutant, Mut2: R1 mutant, Mut3: double R1/R2 mutant) LINC00662 promoters. C Dual luciferase reporter assay was conducted in OS cells transfected with wild-type or mutant reporters and control vector. D Co-localization of PITX1 and STAT3 was detected by immunofluorescence (Scale bars: 20 μm). E Binding of STAT3 and PITX1 was detected by co-IP in HOS and U2OS cells. F Binding of different STAT3 domains and PITX1 were detected by co-IP using HEK 293 T cells. G Expression of STAT3 was detected in PITX1-overexpressing cells following MG132 treatment (10 μm). H Expression of STAT3 was detected by western blotting in control and PITX1-overexpressing OS cells treated with CHX (10 ng/ml) for different times. I Expression of STAT3 was detected by western blotting in control and PITX1-knockdown OS cells treated with CHX (10 ng/ml) for different times. J Immunoprecipitation showing the ubiquitination of STAT3 by ectopic expression of PITX1. STAT3 ubiquitination was detected by immunoprecipitation with anti-STAT3, and then immunoblotted with the indicated antibodies. K Expression of STAT3 and PITX1 in OS and normal tissues was detected by western blotting (n = 4 pairs). Data represent the mean ± SD of three separate determinations. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test