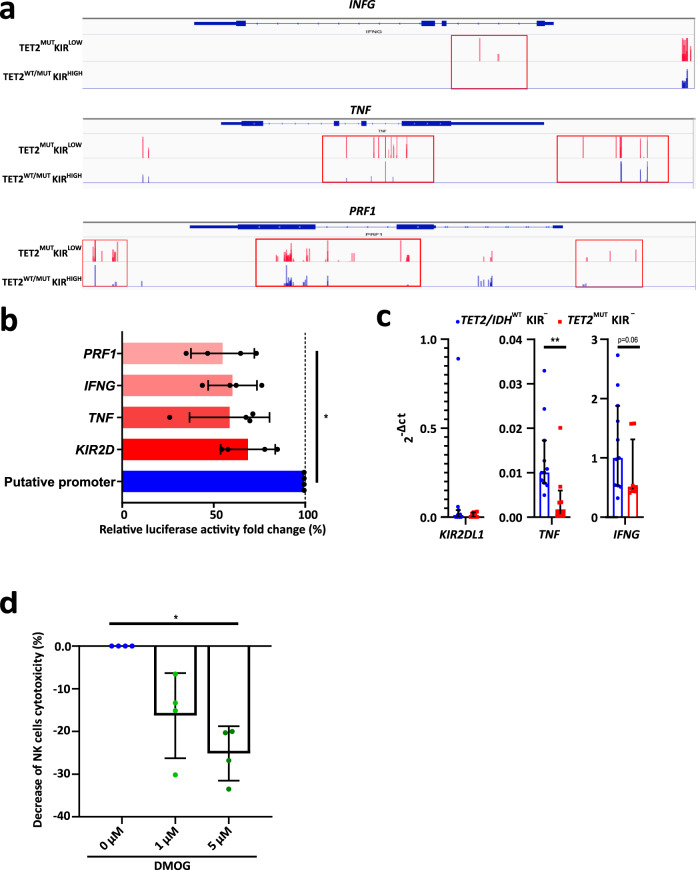
Fig. 5. Hypermethylation of particular CpG sites represses NK cell gene expression and function.
a Methylation profiles, established after RRBS analyses, depicted with IVG at the IFNG (upper panel), TNF (middle panel) and PRF1 (lower panel) loci, were shown in NK cells from patients segregated on the high (TET2WT/MUTKIRHIGH, in blue bars) and low (TET2MUTKIRLOW, in red bars) KIR expression. Red peaks/boxes show significant differences in the DNA methylation levels of these specific loci. b The regulatory activity of specific CpG sites was analyzed in the HEK293T cell line transfected with a luciferase-reporter plasmid including methylated or non-methylated genes’ regulatory regions. Relative luciferase activities of the in vitro methylated regions were compared to their non-methylated counterpart (FC = Methylated/Putative Promoter). Means ± SD is shown (n = 4). Data were analyzed using the one-way Friedman test followed by a Dunn’s test. *p = 0.0433. c Quantification of the KIR2DL1, TNF, and IFNG transcripts by RT-qPCR on KIR2D− NK cells sorted from TET2/IDHWT (n = 11, in blue) and TET2MUT (n = 10, in red) patients. Medians and interquartile are shown. Statistics were calculated with the two-sided Mann–Whitney test, **p = 0.0036. d Fold changes of NK cells killing activity against the NK-sensitive cell line K562 previously treated for 5 days with 1 µM DMOG (n = 10). Each dot represents one independent experiment. DMSO alone was used as a control. Means ± SD is shown. Data were analyzed using the one-way Friedman test followed by a Dunn’s test. *p = 0.0417. Source data are provided as a Source Data file.