Figure 2.
Exercise reprograms transcriptional profiles across tissues in an age-dependent manner
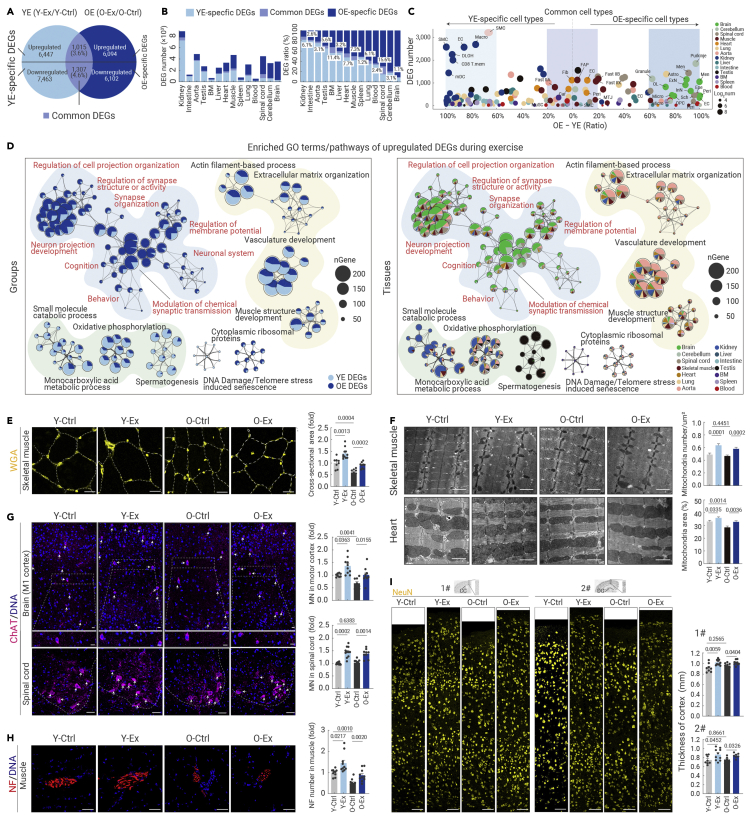
(A) Venn diagram showing the YE-specific DEGs, OE-specific DEGs, as well as common DEGs overlapping between YE DEGs (Y-Ex/Y-Ctrl) and OE DEGs (O-Ex/O-ctrl) based on data from scRNA-seq and snRNA-seq. The DEG numbers were calculated by summing the cell type-specific DEGs from the 14 tissues or organs, yielding the total number of upregulated and downregulated genes.
(B) Bar plots showing the DEG number (left) or DEG relative ratio (corresponding DEG number to total number of DEGs including YE-specific DEGs, common DEGs, and OE-specific DEGs, right) of indicated DEGs across different tissues based on data from scRNA-seq and snRNA-seq.
(C) Bubble plot showing the difference in the DEG ratio of YE DEG number or OE DEG number to total DEG number in different cell types of the 14 tissues based on data from scRNA-seq and snRNA-seq. Cell types that have a DEG ratio difference of more than 60% are named YE-specific cell types or OE-specific cell types. Cell types that have a DEG ratio difference lower than 20% are named Common cell types.
(D) Network plots showing the enriched GO terms/pathways of upregulated DEGs during exercise in the indicated groups (left) or indicated tissues (right) based on data from scRNA-seq and snRNA-seq.
(E) WGA staining of skeletal muscle tissues from the indicated groups. The relative cross-sectional areas are quantified as fold changes. Scale bars, 100 μm. n = 7–12 mice per group.
(F) Electron microscopy analysis of the number of intermyofibrillar mitochondria in skeletal muscle (top) and the percentage of total interfibrillar mitochondrial area in heart tissues (bottom) from the indicated groups. Scale bars, 1 μm. n = 4 mice per group and 10 images were taken from each mouse.
(G) Immunofluorescence analysis of ChAT-positive motor neurons (MNs) in the motor cortex and spinal cord from the indicated groups. Scale bars, 50 μm. n = 7–12 mice per group.
(H) Immunofluorescence analysis of neurofilament (NF)-positive terminal buttons in skeletal muscle tissues from the indicated groups. The number of terminal buttons per 0.6 mm2 is measured across different groups. Scale bars, 50 μm. n = 7–12 mice per group.
(I) Immunochemistry analysis of NeuN-positive neurons in the brain cortex from the indicated groups. The thickness of the cortex from the indicated regions was measured across different groups. Scale bars, 50 μm. n = 7–12 mice per group. 1# and 2# cortexes are the motor and sensory cortex above the corpus callosum (CC) and dentate gyrus (DG) regions, respectively. The quantification data in (F)–(I) are shown as the means ± SEM, and two-tailed Student's t test p values are indicated.