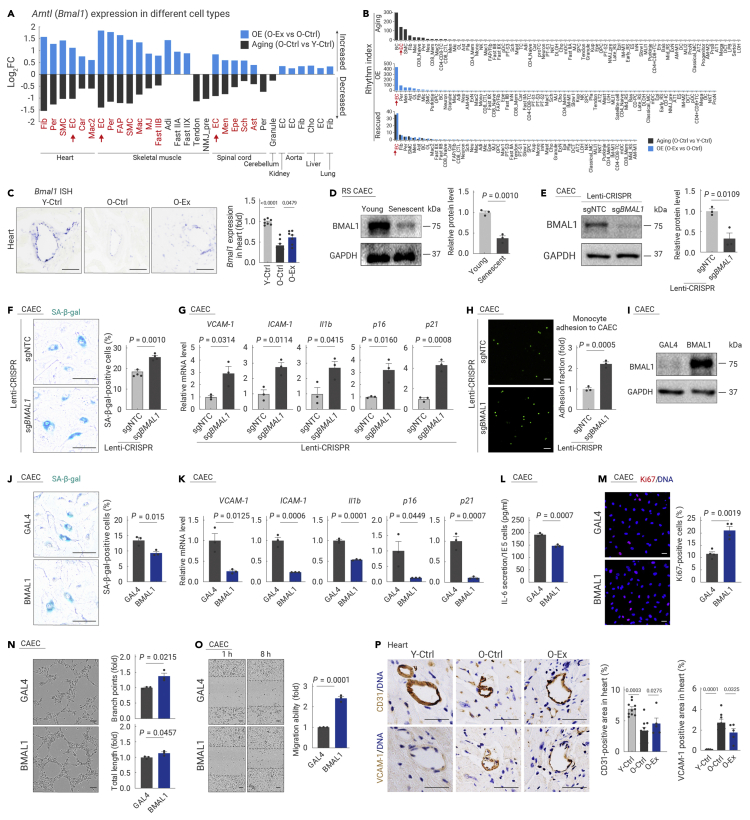
Figure 7.
BMAL1 plays a protective role in endothelial cells
(A) Bar plot showing the expression changes of Arntl (Bmal1) in different cell types from all the tissues analyzed in the indicated groups.
(B) Bar plots showing the rhythm index of different cell types from all the tissues analyzed in the indicated groups.
(C) Transverse sections of heart tissues from the indicated groups of mice were subjected to RNA in situ hybridization (RNA-ISH) with Bmal1 riboprobes. Left, representative ISH images; right, the relative expression level of Bmal1 in mouse hearts was quantified. Scale bars, 50 μm. n = 7 mice per group.
(D) Western blotting of BMAL1 protein levels in CAECs at the indicated passages. n = 3 biological repeats per group. Young, passage 4; Senescent, passage 11. RS, replicative senescence.
(E) Western blotting of BMAL1 protein levels in CAECs transduced with lentivirus with non-targeting or Bmal1-targeting sgRNAs in the CRISPR-Cas9-mediated knockout system.
(F) SA-β-gal analysis of CAECs transduced with lentivirus with non-targeting or Bmal1-targeting sgRNAs in the CRISPR-Cas9-mediated knockout system. Scale bars, 100 μm. n = 4 biological repeats per group.
(G) Transcript levels of indicated genes in CAECs transduced with lentivirus with non-targeting or Bmal1-targeting sgRNAs in the CRISPR-Cas9-mediated knockout system. n = 3 biological repeats per group.
(H) Monocyte adhesion assay in CAECs transduced with lentivirus with non-targeting or Bmal1-targeting sgRNAs in the CRISPR-Cas9-mediated knockout system. Scale bars, 50 μm. n = 3 biological repeats per group.
(I) Western blotting of BMAL1 protein levels in CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. n = 3 biological repeats per group.
(J) SA-β-gal analysis of CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. The percentage of SA-β-gal-positive cells was quantified. Scale bars, 100 μm. n = 3 biological repeats per group.
(K) Transcript levels of the indicated genes associated with endothelial cell dysfunction in CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. n = 3 biological repeats per group.
(L) ELISA analysis of the secretion of IL-6 in CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. The absorption value is normalized to the cell number. n = 3 biological repeats per group.
(M) Immunofluorescence staining of Ki67 in CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. Scale bars, 50 μm. n = 4 biological repeats per group.
(N) Assessment of tube formation capability in CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. The relative branch points (right upper) and total length of tubes (right lower) were quantified as fold changes. Scale bar, 100 μm. n = 3 biological repeats per group.
(O) Wound scratch assay showing the migration ability of CAECs transduced with lentivirus-expressing Flag-tagged GAL4 or BMAL1. Red lines represent scar boundaries. The relative migration ability was quantified as the fold change. Scale bar, 100 μm. n = 3 biological repeats per group.
(P) Immunochemistry analysis of CD31-positive (left) and VCAM-1-positive (right) cells in the heart using adjacent tissue sections from the indicated groups. The percentages of CD31- or VCAM-1-positive areas were quantified. Scale bars, 50 μm. n = 5–12 mice per group.
The quantification data in (C)–(P) are shown as the means ± SEM, and two-tailed Student's t test p values are indicated.