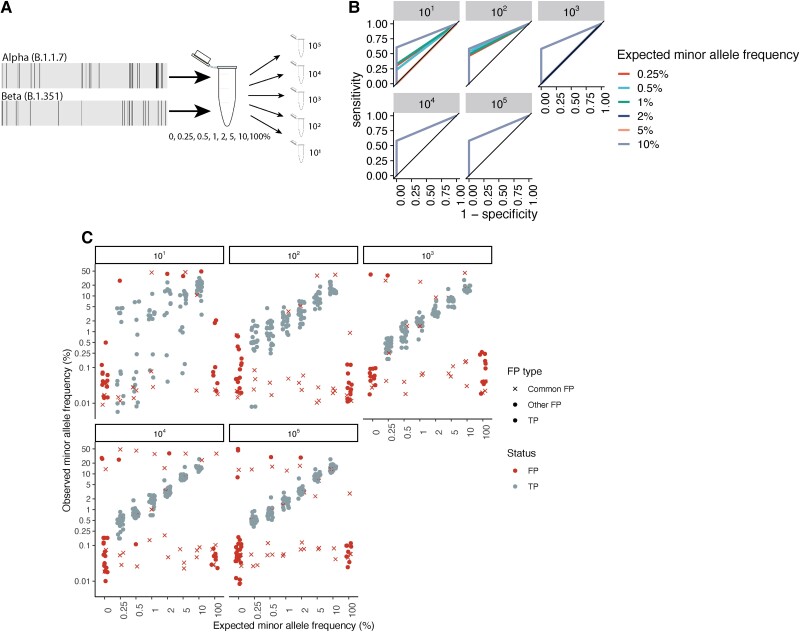
Figure 2.
Measuring the accuracy of within-host severe acute respiratory syndrome coronavirus 2 variant identification. (A) Diagram of artificial strain mixture experiment. We conducted a serial dilution experiment, mixing synthetic ribonucleic acid (RNA) controls (Twist Biosciences) of the Alpha (B.1.1.7) and Beta (B.1.351) variants so that the minor variant comprised 0%–10% of the source material and then serially diluted mixtures to a total of 101–105 total RNA copies. We conducted amplicon-based sequencing of artificial mixtures, identified intrahost single-nucleotide variants (iSNVs) with the viralrecon pipeline [24], and determined the sensitivity and specificity of our variant calling pipeline to recovering true variation within our synthetic mixtures. (B) Receiver operator characteristic curve showing 1 specificity versus sensitivity in the recovery of true minority variants, colored by total RNA dilution. Lines corresponding to each dilution include results from 5 artificial strain mixtures, including minority variants present at 0.5%–10% of the total viral pool. (C) Observed minor allele frequency versus expected minor allele frequency for artificial strain mixtures. Points indicate iSNV assignment (false-positive [FP] iSNV; true positive [TP] iSNV). For FP iSNVs, point shape indicates whether FPs were commonly repeated across samples (common FP, FP identified in 10 or more samples; other FP, any other FP iSNV). Points were jittered for visualization. Horizontal facets indicate the synthetic RNA copy number in units of genome copies per microliter.