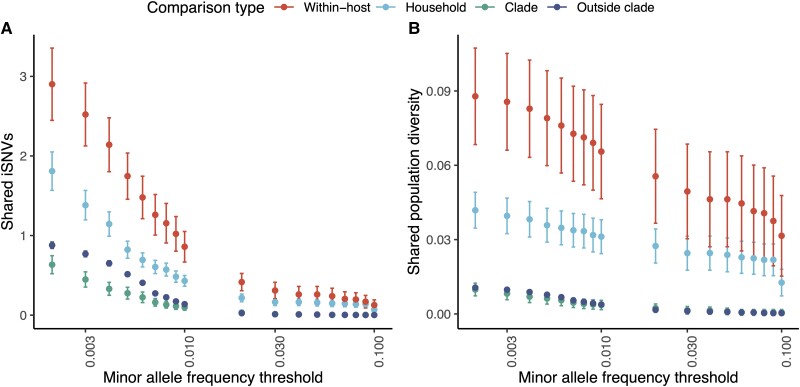
Figure 3.
Shared within-host variants hold a signal of severe acute respiratory syndrome coronavirus 2 transmission. For our genomic collection from a household transmission study in California, (A) pairwise comparisons of the number of shared intrahost single-nucleotide variants (iSNVs), defined as a shared minor allele present at the same genomic position, and (B) shared population diversity, as measured by the geometric mean of the sum of within-host minor allele frequencies for shared iSNVs, identified across different minor allele frequency thresholds, after applying all variant filters (see Methods). Points and error bars indicate mean and 95% confidence intervals and are colored by comparison type. Each pair is assigned to a unique category. Within-host, pairs of samples from the same individual collected on different days; Household, pairs of individuals from the same household; Clade, pairs of individuals outside households infected with the same Nextclade clade; and Outside clade, pairs of individuals outside households infected with different Nextclade clades. Pairwise comparisons include only samples sequenced in different sequencing batches.