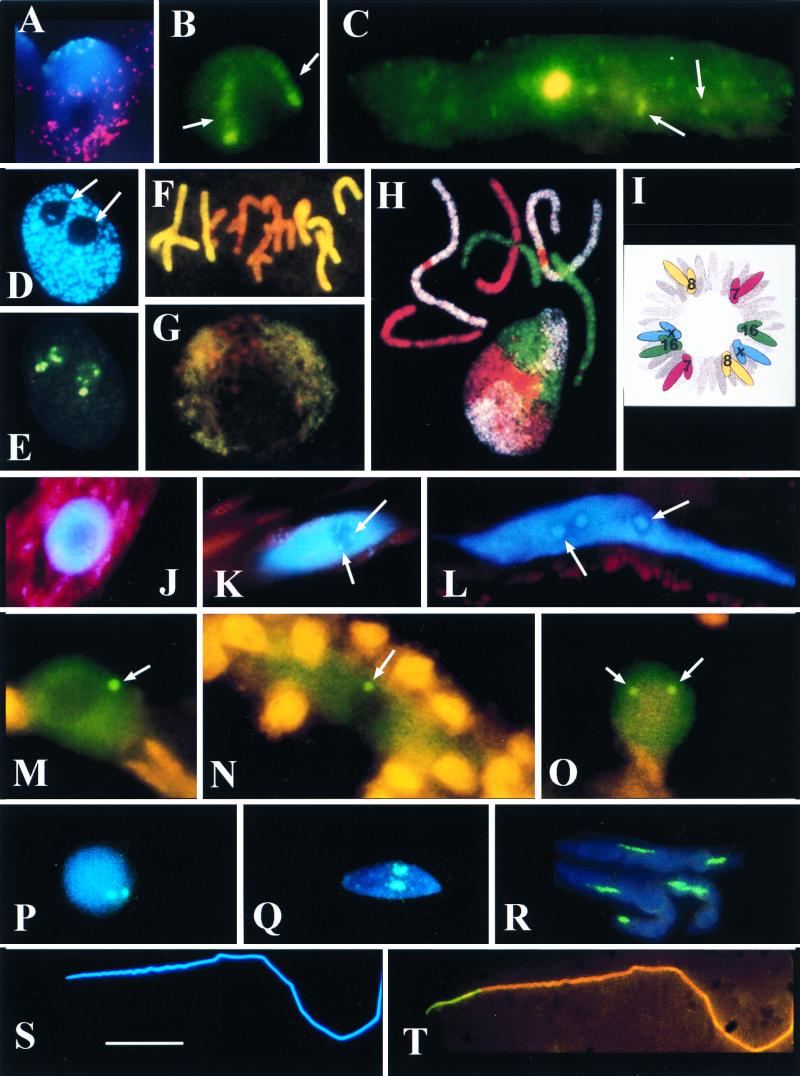
FIG. 1.
(A and B) Triticum aestivum (wheat) root tip meristematic nuclei. (A) CCS1 labelling for centromeres (digoxigenin-fluorescein isothiocyanate [FITC], cyan fluorescence) at one pole and the telomere consensus sequence (TTTAGGG)n, biotin-Cy3, red fluorescence) at the other. The red dots apparently outside the central nucleus correspond to signal from the telomeric pole of an adjacent nucleus. The nucleus is counterstained for DNA (DAPI stain, blue fluorescence). Photo courtesy of L. Aragon-Alacaide and G. Moore. (B) T. aestivum cv. Beaver carrying two 1Bl/1Rs chromosome arms has 1Rs detected by GISH (digoxigenin-labelled total Secale cereale [rye] DNA-FITC, green fluorescence). Note that the two elongate 1Rs chromosome arm domains (arrows) with the condensed subtelomeric heterochromatin fluoresce more strongly than the remainder of the arm. (C) Protophloem nucleus of T. aestivum cv. Beaver labelled by GISH with total rye DNA (digoxigenin-FITC, green fluorescence). Note that the nucleus is much larger and more elongated than in panel B and is endoreduplicated; the single large 1Rs domain is in the center of the nucleus; there is no evidence of elongate chromosome domains; and fragments of rye signal across the whole volume of the nucleus (arrows). (D and E) Sectioned nucleus from a wheat meristematic cell. (D) The nucleus is counterstained with DAPI (blue fluorescence) for DNA. Note the two spherical nucleoli (arrows). (E) The same nucleus section labelled for rDNA (digoxigenin-labelled pTa71 [59]-FITC, yellow fluorescence). The rDNA signal occurs outside the nucleolus (arrowed in panel D) on a condensed chromatin fiber and inside the nucleolus on chromatin fibers with different levels of condensation (compare with panel D). (F and G) Root tip meristematic metaphase (F) and interphase (G) of the hybrid Hordeum vulgare (barley) × Secale africanum (wild rye) labelled by GISH with total DNA from the wild rye parent (digoxigenin-FITC, yellow fluorescence) and counterstained with propidium iodide for DNA (orange fluorescence). (F) The metaphase plate shows genome separation, with seven chromosomes of wild-rye origin at the periphery and the seven chromosomes of barley origin at the center. (G) Genome separation at interphase with wild-rye chromatin outside the central barley genome. Panels F and G are taken from reference 101. (H) Chromosome painting of a metaphase and interphase nucleus of female fibroblasts of Muntiacus muntjac vaginalis (Indian munjac), chromosome 1 (biotin-Cy5, white), chromosome 2 (FITC–12-dUTP, green), chromosome X +3 (Cy3-dUTP, red) (157). Note that the chromosomes occur in discrete unpaired domains at interphase. Photograph courtesy of F. Yang and M. Ferguson-Smith. Taken from Chromosome Research. (I) Diagram of human fibroblast prometaphase showing the positions of chromosomes 7, 8, 16, and X (from reference 120). Note that a complete set of identified chromosomes are drawn on each side of the prometaphase, suggesting genome separation. Nagele et al. (120) also suggest that there is an order of chromosomes in each genome (i.e. 7, 16, X, 8) in two antiparallel sets. (J to O) Funaria hygrometrica (moss) nuclei from caulonemata (J to N) and from a thallus cell (O). (J to L) DAPI-stained nuclei (blue fluorescence) from an apical cell (J), cell 8 (K), and cell 15 (L) of the caulonemata filament. Note the increasing size and elongation of nucleus and the accumulation of rDNA heterochromatin (arrows). (M to O) Immunocytochemistry to detect D-polypeptide of the spliceosome complex (FITC detection, green fluorescence). In addition to a uniform dispersal of signal across the nucleus but outside the nucleolus, there is one coiled body in the nuclei of cells 4 (M) and 10 (N) of caulonema and two coiled bodies in the nuclei of thallus cells (arrows). All coiled bodies are associated with the nucleolus (O). (P to T) Spermatogenesis in Schistocerca gregaria (locust) stained blue for DNA with DAPI (blue fluorescence) and labelled for rDNA (pTa71, digoxigenin-FITC, green/cyan fluorescence). (P to R) Detection of rDNA in double exposures with DAPI (blue/cyan fluorescence). (P) Early spermatid nucleus with two rDNA loci. (Q and R) Increasingly mature and elongated spermatid nuclei. Note the elongating rDNA loci. (S and T) Fully mature and elongated spermatozoan nucleus, DAPI stained (S) and probed for rDNA (T, yellow fluorescence). Note that all the rDNA signal (yellow) is basal to the nucleus, suggesting intranuclear migration of rDNA. Scale bars, 15 μm for panels A, B, C, F, G, M, N, O, P, Q, R, S, and T; 10 μm for panels D and E; and 20 μm for panels H, J, K, and L.