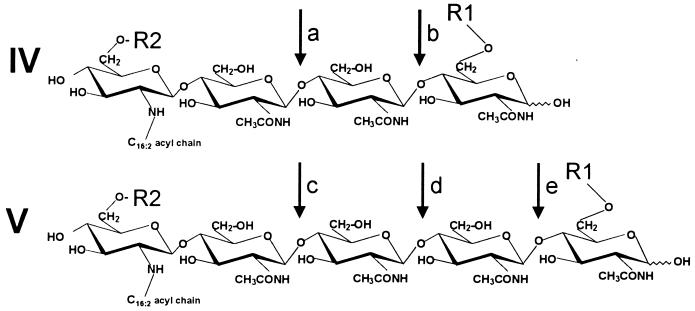
TABLE 7.
Sites of hydrolytic cleavage of Nod factors of Rhizobium meliloti by enzymes isolated from the roots of Medicago sativaa
| Hydrolase (reference) | No. of N-acetylglucosamine residues | R1 | R2 | Cleavage site(s) |
|---|---|---|---|---|
| Intact roots (258, 260) | IV | Sulfatyl | H | a |
| IV | H | H | a, b | |
| IV | Sulfatyl | O-Acetyl | a | |
| V | Sulfatyl | H | c, d | |
| V | H | H | c, d, e | |
| Chitinase class I (241, 258) | IV | Sulfatyl | H | None |
| IV | H | H | b | |
| IV | Sulfatyl | O-Acetyl | None | |
| V | Sulfatyl | H | d | |
| V | H | H | d, e | |
| Novel chitinase/lysozyme (179) | IV | Sulfatyl | H | b |
| IV | H | H | b | |
| IV | Sulfatyl | O-Acetyl | b | |
| V | Sulfatyl | H | d | |
| Nod factor hydrolase (258, 260) | IV | Sulfatyl | H | a |
| IV | H | H | a | |
| IV | Sulfatyl | O-Acetyl | a | |
| V | Sulfatyl | H | c | |
| V | H | H | c | |
| V | Sulfatyl | O-Acetyl | c |
Distinct cleavage site preferences exist toward tetrameric (IV) and pentameric (V) Nod factors of R. meliloti as well as their desulfated and deacetylated derivatives, respectively.