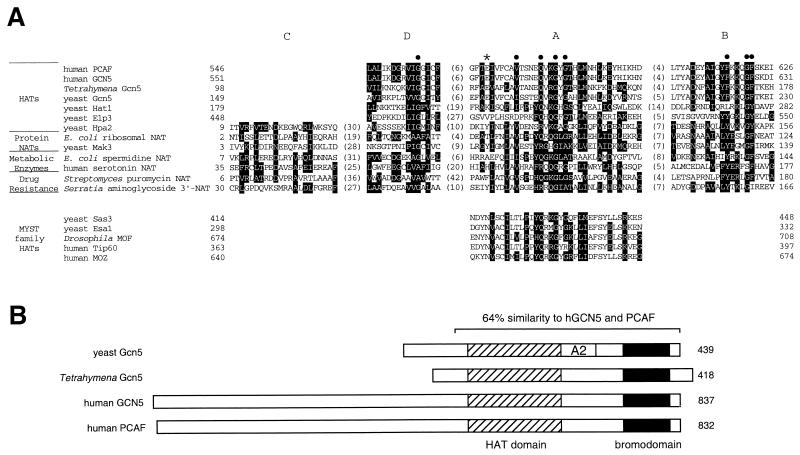
FIG. 1.
Similarities of GNAT (Gcn5-related N-acetyltransferase) superfamily members. (A) Alignment of GNAT homology motifs A, B, C, and D for HATs and representatives of other types of acetyltransferases. Reversed type indicates consensus sequence residues, as determined by Neuwald and Landsman (174), and solid circles mark residues that are particularly conserved throughout the superfamily. The asterisk indicates the glutamate residue known to be critical for HAT catalysis in yeast Gcn5. At the bottom are several members of another acetyltransferase family, the MYST proteins, that share just the A motif. (B) Alignment of proteins from the Gcn5 subgroup of the GNAT superfamily, showing the location of the HAT domain and bromodomain. Shown at the top is the overall homology region shared by all four proteins, with the similarity between the yeast and human proteins indicated; Tetrahymena Gcn5 has 62% similarity with the three others over this same region. The A2 label designates a region in yeast Gcn5 known to interact with the adaptor protein Ada2 (37). In addition, the N-terminal region of PCAF has been generally defined as its site of interaction with p300/CBP and nuclear receptor coactivator SRC-1 (130, 276); this region and the HAT domain also interact with viral protein E1A (40, 193).