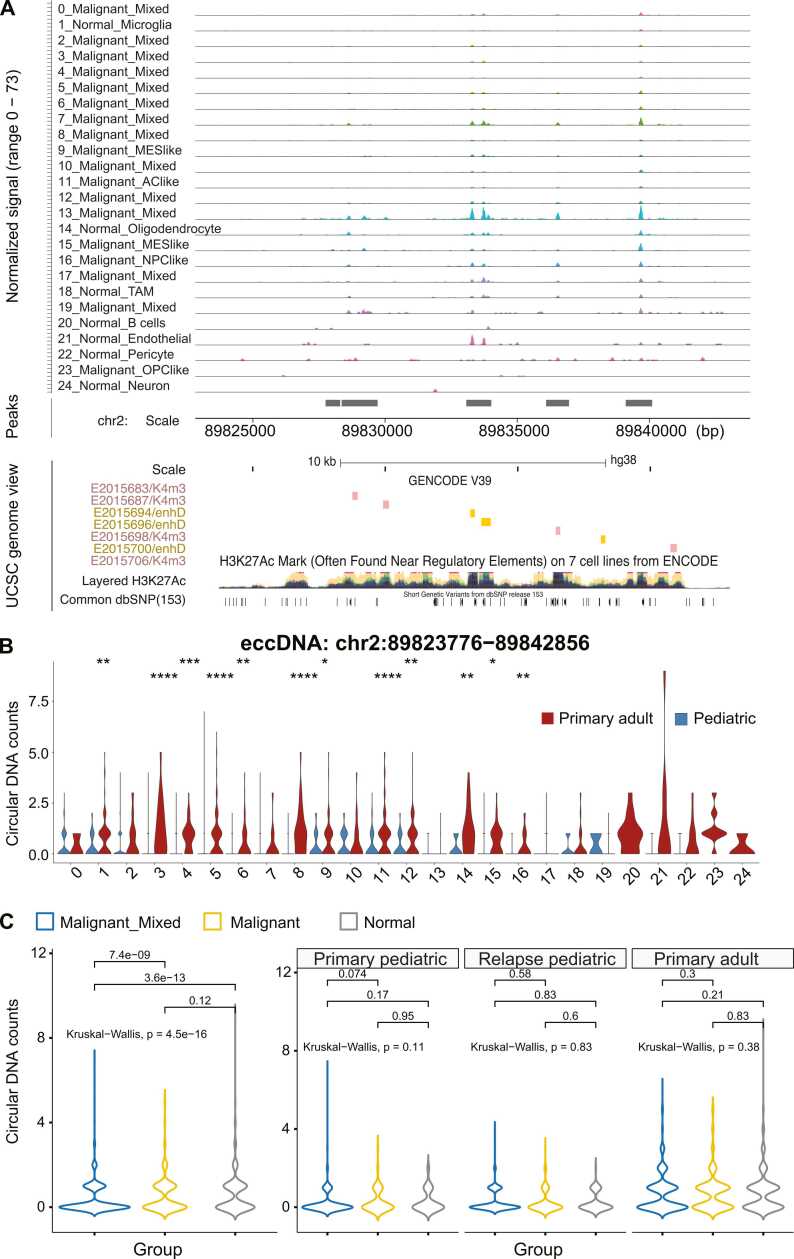
Fig. 2.
Single-cell analysis revealed cell type-specific coverage of eccDNA chr2:89823776−89842856. (A) Linear read coverage view (top track; each row represents a Seurat cell cluster) for eccDNA chr2:89823776−89842856, together with track view of peaks (middle) and UCSC Genome Browser view (bottom). Overlapping regulatory elements are colored by annotation sources, together with layered H3K27ac histone signal for enhancers shown in the bottom track. Abbreviations: TAM: tumor-associated macrophage; NPClike, neural-progenitor-like; OPClike, oligodendrocyte-progenitor-like; AClike, astrocyte-like; MESlike, mesenchymal-like. (B) Circular coverage of eccDNA at locus chr2:89823776−89842856 in GBM samples. Seurat cell clusters are named in the format of [Cluster ID]_[Tumor or normal cell]_[cell type or cell states]. Mixed: the malignant cluster is a mixture of at least two cell states out of MESlike, OPClike, NPClike and AClike. The non-parametric Wilcox test was applied to compare eccDNA distributions by cell type between adult and pediatric conditions. Significance: ****: p < 0.0001. ***: p < 0.001. **: p < 0.01. *: p < 0.05. (C) Comparison of eccDNA chr2:89823776−89842856 between malignant and normal cells. Left penal: comparison of the eccDNA distribution across groups of malignant, malignant_mixed and normal cells. Right penal: the left penal comparison was further stratified by tumor sample condition. The non-parametric Kruskal-Wallis test was used to test whether the three cell groups were originated from the same distribution.