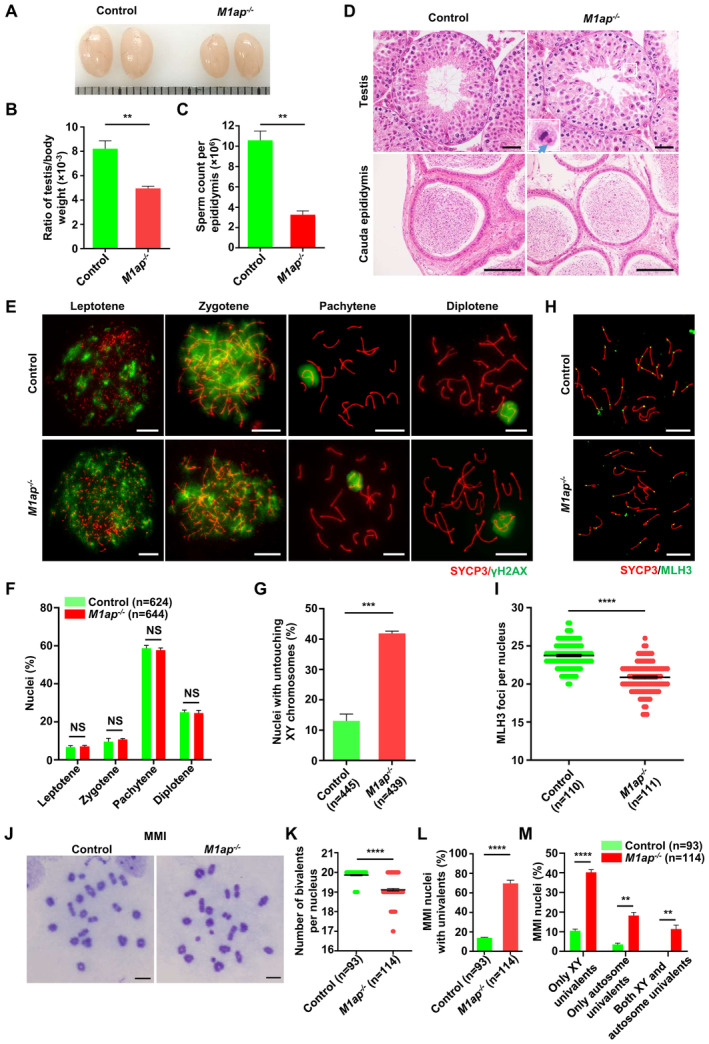
Figure EV4. The phenotype of M1ap −/− mice.
-
A–CRepresentative images of testes (A), the ratio of testis/body weight (B), and the sperm count per epididymis (C) from 8‐week‐old control and M1ap −/− mice. Each grid represents 1 mm in (A).
-
DRepresentative images of hematoxylin and eosin‐stained sections of testes and cauda epididymides from 8‐week‐old control and M1ap −/− mice. A magnified view of the boxed representative metaphase cell is shown in the lower left corner of the image of M1ap −/− mice. The blue arrow indicates unaligned chromosomes. Scale bars, 50 μm.
-
EImmunofluorescence staining of SYCP3 (red) and γH2AX (green) on spermatocyte spreads from 8‐week‐old control and M1ap −/− mice. Scale bars, 10 μm.
-
FThe percentages of spermatocytes at each stage of meiotic prophase I in 8‐week‐old control and M1ap −/− mice.
-
GQuantification of nuclei with untouching XY chromosomes at pachytene.
-
HImmunofluorescence staining of SYCP3 (red) and MLH3 (green) in spermatocyte spreads from 8‐week‐old control and M1ap −/− mice. Scale bars, 10 μm.
-
INumber of MLH3 foci per cell.
-
JMeiotic metaphase I (MMI) spermatocytes stained with Giemsa. Scale bars, 10 μm.
-
KNumber of bivalents per nucleus.
-
LFrequencies of nuclei with univalents.
-
MFrequencies of nuclei with only XY univalents, only autosome univalents, or both XY and autosome univalents.
Data information: (B and C), Data are presented as the mean ± SEM of three biological replicates. (F, G, I, and K–M) Data are presented as mean ± SEM. n shows the number of cells scored from at least three biological replicates. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant; two‐tailed Student's t‐test.
