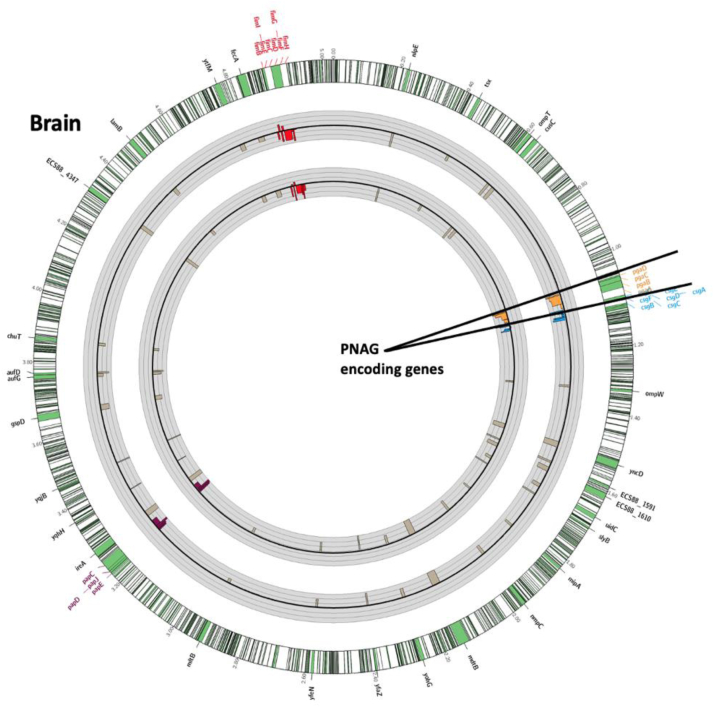
Fig. 3.
In vivo (brain infection) genetic fitness analysis using TnSeq of E. coli K1 mutants in genes encoding extracellular products or outer-membrane proteins. The outermost circle represents the full E. coli K1 S88 genome with a 30-times magnification of the regions of interest shown as green portions of the chromosome. The first gray circle represents the fold change of the reads of the Tn insertions from the Liver to the Brain after gavage. The second gray circle represents the fold change of the reads of the Tn insertions from the LB to the Brain after gavage. Black circular lines in the middle of the grey circles represent baseline. Thin grey circular lines above and below the central black circular line represent 10-fold-changes (i.e., a log10 scale). The limit of the fold change representation is 1000. Bars represent changes in individual genes. Bars pointing toward the circle's center represent Tn interrupted genes resulting in negative fitness whereas bars pointed outward represent genes whose loss increases fitness. Poly-β-(1-6)-N-Acetyl-Glucosamine (PNAG), Curly, Type 1 Fimbriae and Type 1 pilus encoding genes are highlighted in red, purple, blue and light brown respectively.