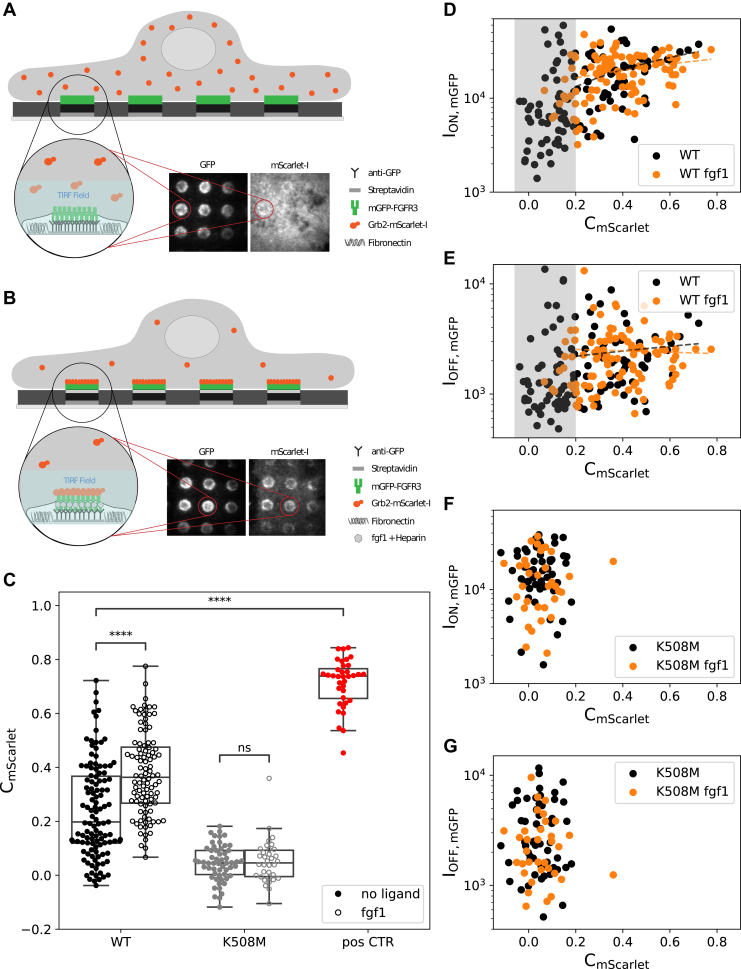
Figure 1.
Experimental design and proof of principle.A and B, antibody patterns are used to enrich and immobilize mGFP-FGFR3 at specific sites (“ON” regions) in the plasma membrane of HeLa cells, leaving other regions depleted of mGFP-FGFR3 (“OFF”). Colocalization of the adaptor protein GRB2-mScarlet to mGFP-FGFR3 patterns reports on the activation state of FGFR3, with no or little copatterning observable in the nonactivated state (A) and a high degree of copatterning for the activated receptor after addition of the ligand fgf1 (B). TIRF illumination is used to specifically detect membrane-proximal protein. C, the fluorescence contrast of GRB2-mScarlet (CmScarlet) relates the fluorescence intensity within ON (ION,mGFP) and OFF (IOFF,mGFP) areas of FGFR3-enriched regions and serves to quantify the extent of colocalization. Each dot represents one cell. CmScarlet data for the WT receptor, a kinase-dead mutant (K508M) and an mGFP-FGFR3-mScarlet fusion protein as positive control is shown (p value annotation legend: ∗0.01 ≤ p ≤ 0.05; ∗∗0.001 ≤ p ≤ 0.01; ∗∗∗0.0001 ≤ p ≤ 0.001; and ∗∗∗∗p ≤ 0.0001). D and E, correlation between the receptor’s intensity in ON (D) and OFF (E) regions and the GRB2-mScarlet contrast for the WT receptor. Data in the absence (black) and presence (orange) of fgf1 are shown. The gray box indicates the cell population with CmScarlet <0.2, which likely represents nonactivated cells. F and G, correlation between GRB2-mScarlet contrast and mGFP-FGFR3 intensity in ON (F) and OFF (G) regions for K508M. All correlation coefficients can be found in Table S1. FGFR3, fibroblast growth factor receptor 3; GRB2, growth factor receptor–bound 2; mGFP, monomeric GFP; TIRF, total internal reflection fluorescence.